Today, give a try to Techtonique web app, a tool designed to help you make informed, data-driven decisions using Mathematics, Statistics, Machine Learning, and Data Visualization. Here is a tutorial with audio, video, code, and slides: https://moudiki2.gumroad.com/l/nrhgb. 100 API requests are now (and forever) offered to every user every month, no matter the pricing tier.
Survival analysis is a group of Statistical/Machine Learning (ML) methods for predicting the time until an event of interest occurs. Examples of events include (link to notebook at the end):
- death
- failure
- recovery
- default
- etc.
And the event of interest can be anything that has a duration:
- the time until a machine breaks down
- the time until a customer buys a product
- the time until a patient dies
- etc.
The event can be censored, meaning that it has’nt occurred for some subjects at the time of analysis.
In this post, I show how to use scikit-learn, glmnet, xgboost, lightgbm, pytorch, keras, nnetsauce and mlsauce in conjuction with Python package survivalist for probabilistic survival analysis. The probabilistic part is based on conformal prediction and Bayesian inference, and graphics represent the out-of-sample ML survival function vs Empirical Kaplan-Meier survival function (with confidence intervals).
A link to the corresponding notebook can be found at the end of this post.
Contents
- Contents
- 0 - Installation
- 1 - using
scikit-learnwith conformal prediction - 2 - using
nnetsauce - 3 - using
glmnet - 4 - using
pytorch - 5 - Using keras (through
scikeras) - 6 - using
xgboost - 7 - using
lightgbm - 8 - using Generic Boosting (
mlsauce)
0 - Installation
!pip uninstall -y survivalist
!pip install survivalist --upgrade --no-cache-dir
!pip install glmnetforpython --verbose --upgrade --no-cache-dir
!pip install nnetsauce --verbose --upgrade --no-cache-dir
!pip install scikeras
!pip install xgboost --upgrade --no-cache-dir
!pip install lightgbm --upgrade --no-cache-dir
!pip install git+https://github.com/Techtonique/mlsauce.git --verbose
import numpy as np
import pandas as pd
def _encode_categorical_columns(df, categorical_columns=None):
"""
Automatically identifies categorical columns and applies one-hot encoding.
Parameters:
- df (pd.DataFrame): The input DataFrame with mixed continuous and categorical variables.
- categorical_columns (list): Optional list of column names to treat as categorical.
Returns:
- pd.DataFrame: A new DataFrame with one-hot encoded categorical columns.
"""
# Automatically identify categorical columns if not provided
if categorical_columns is None:
categorical_columns = df.select_dtypes(include=['object', 'category']).columns.tolist()
# Apply one-hot encoding to the identified categorical columns
df_encoded = pd.get_dummies(df, columns=categorical_columns)
# Convert boolean columns to integer (0 and 1)
bool_columns = df_encoded.select_dtypes(include=['bool']).columns.tolist()
df_encoded[bool_columns] = df_encoded[bool_columns].astype(int)
return df_encoded
import matplotlib.pyplot as plt
import nnetsauce as ns
import glmnetforpython as glmnet
from survivalist.nonparametric import kaplan_meier_estimator
from survivalist.datasets import load_whas500, load_gbsg2, load_veterans_lung_cancer
from survivalist.ensemble import ComponentwiseGenGradientBoostingSurvivalAnalysis
from survivalist.custom import SurvivalCustom
from survivalist.custom import PISurvivalCustom
from survivalist.ensemble import GradientBoostingSurvivalAnalysis
from sklearn.linear_model import RidgeCV, ElasticNetCV
from sklearn.ensemble import RandomForestRegressor
from sklearn.tree import ExtraTreeRegressor
from sklearn.kernel_ridge import KernelRidge
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from survivalist.ensemble import PIComponentwiseGenGradientBoostingSurvivalAnalysis
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split
from time import time
import matplotlib.pyplot as plt
import nnetsauce as ns
import numpy as np
from survivalist.datasets import load_whas500, load_veterans_lung_cancer, load_gbsg2
from survivalist.custom import SurvivalCustom
from sklearn.linear_model import BayesianRidge, ARDRegression
from sklearn.gaussian_process import GaussianProcessRegressor
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestRegressor, ExtraTreesRegressor
from sklearn.neural_network import MLPRegressor
from survivalist.metrics import brier_score, integrated_brier_score
from time import time
import pandas as pd
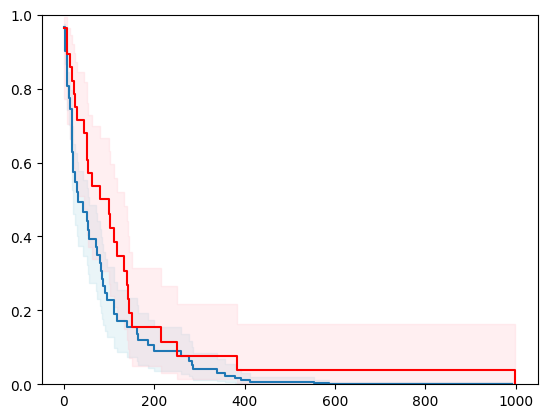
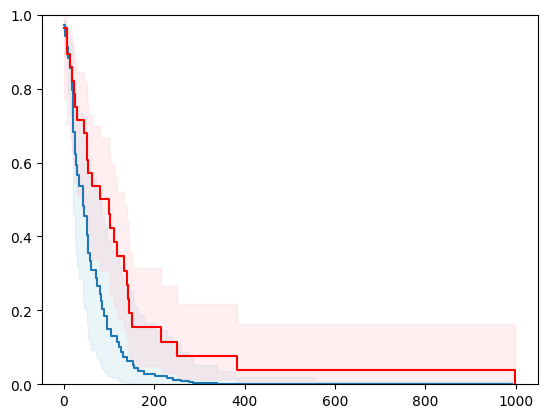
1 - using scikit-learn with conformal prediction
X, y = load_whas500()
X = _encode_categorical_columns(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PIComponentwiseGenGradientBoostingSurvivalAnalysis(regr=RidgeCV(), type_pi="bootstrap")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
100%|██████████| 100/100 [00:09<00:00, 10.54it/s]
100%|██████████| 100/100 [00:10<00:00, 9.61it/s]
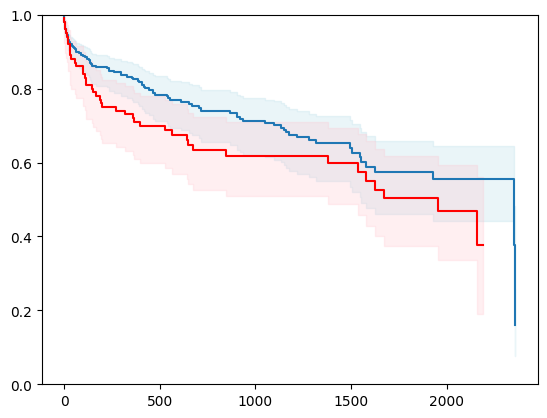
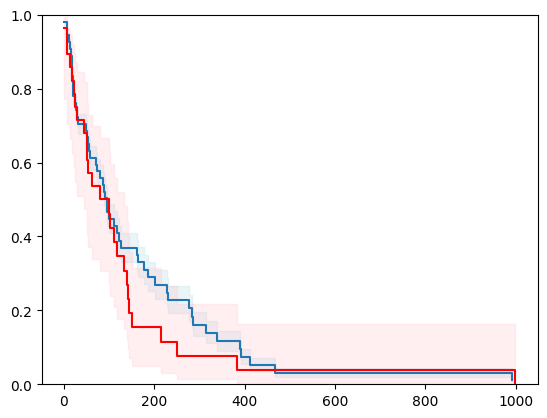
X, y = load_gbsg2()
X = _encode_categorical_columns(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PIComponentwiseGenGradientBoostingSurvivalAnalysis(regr=RidgeCV(), type_pi="kde")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
100%|██████████| 100/100 [00:02<00:00, 42.51it/s]
100%|██████████| 100/100 [00:02<00:00, 42.67it/s]
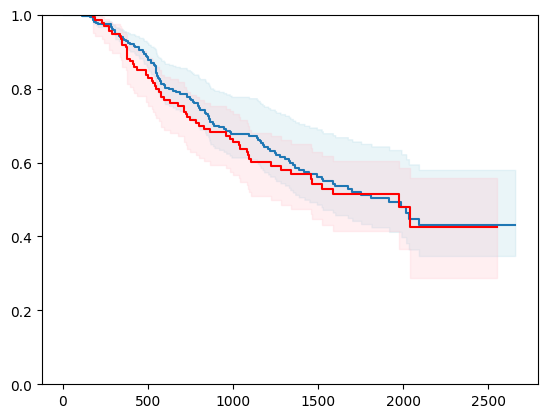
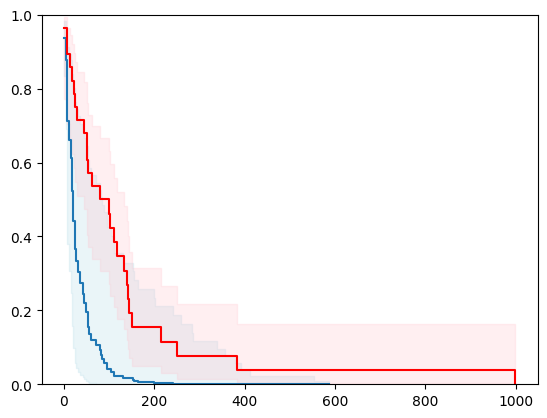
X, y = load_veterans_lung_cancer()
X = _encode_categorical_columns(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PIComponentwiseGenGradientBoostingSurvivalAnalysis(regr=RidgeCV(), type_pi="kde")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
100%|██████████| 100/100 [00:02<00:00, 44.45it/s]
100%|██████████| 100/100 [00:02<00:00, 35.99it/s]
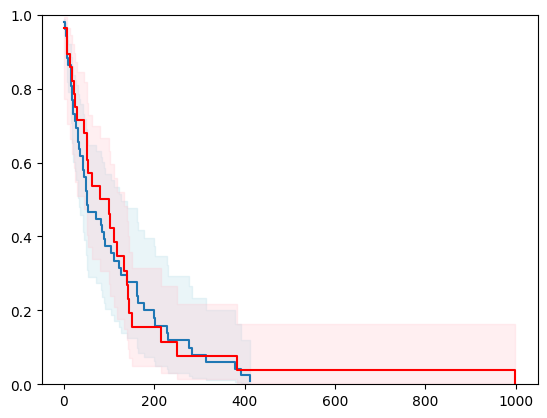
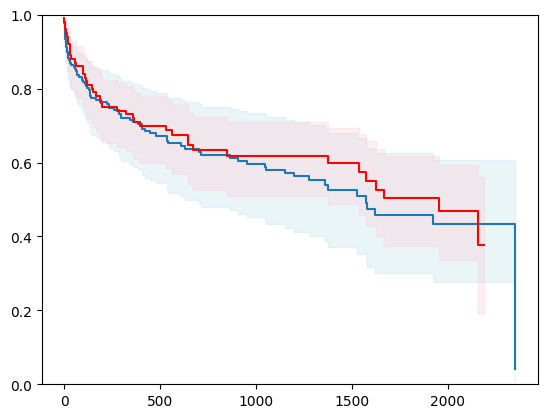
X, y = load_veterans_lung_cancer()
X = _encode_categorical_columns(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PIComponentwiseGenGradientBoostingSurvivalAnalysis(regr=RidgeCV(), type_pi="ecdf")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
100%|██████████| 100/100 [00:01<00:00, 51.44it/s]
100%|██████████| 100/100 [00:01<00:00, 51.30it/s]
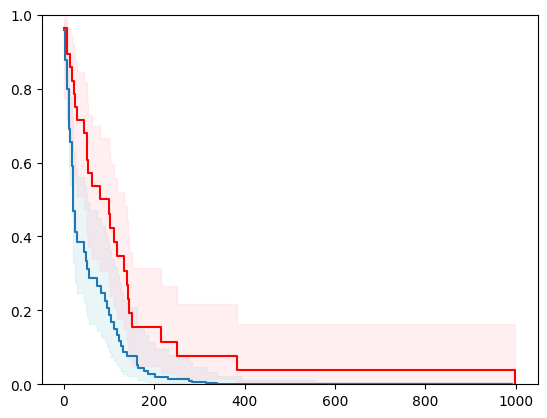
X, y = load_veterans_lung_cancer()
X = _encode_categorical_columns(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PIComponentwiseGenGradientBoostingSurvivalAnalysis(regr=RidgeCV(), type_pi="kde")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
100%|██████████| 100/100 [00:01<00:00, 51.20it/s]
100%|██████████| 100/100 [00:01<00:00, 51.52it/s]
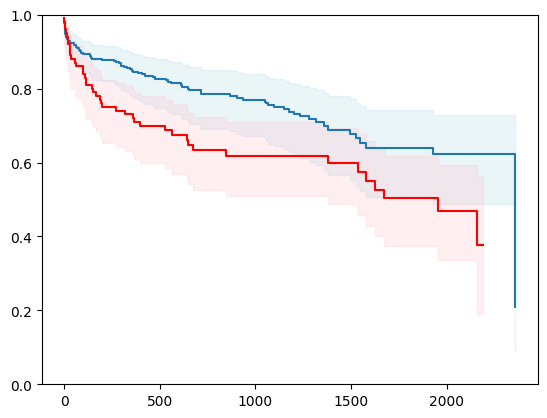
2 - using nnetsauce
2 - 1 with conformal prediction
X, y = load_whas500()
X = _encode_categorical_columns(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PIComponentwiseGenGradientBoostingSurvivalAnalysis(regr=ns.CustomRegressor(RidgeCV()),
type_pi="bootstrap")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
100%|██████████| 100/100 [00:42<00:00, 2.35it/s]
100%|██████████| 100/100 [00:40<00:00, 2.46it/s]
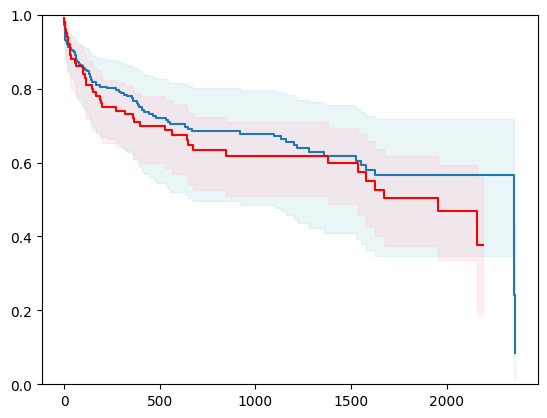
from pickle import Pickler
X, y = load_whas500()
X = _encode_categorical_columns(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PISurvivalCustom(regr=ns.CustomRegressor(RidgeCV()), type_pi="kde")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
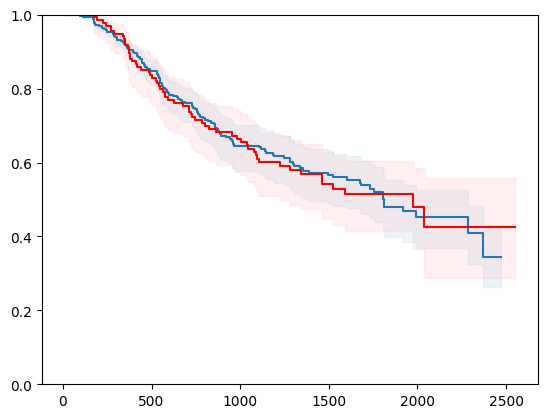
X, y = load_gbsg2()
X = _encode_categorical_columns(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PIComponentwiseGenGradientBoostingSurvivalAnalysis(regr=ns.CustomRegressor(RidgeCV()),
type_pi="kde")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
100%|██████████| 100/100 [00:22<00:00, 4.48it/s]
100%|██████████| 100/100 [00:21<00:00, 4.71it/s]
X, y = load_veterans_lung_cancer()
X = _encode_categorical_columns(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PIComponentwiseGenGradientBoostingSurvivalAnalysis(regr=ns.CustomRegressor(RidgeCV()),
type_pi="kde")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
100%|██████████| 100/100 [00:17<00:00, 5.88it/s]
100%|██████████| 100/100 [00:18<00:00, 5.42it/s]
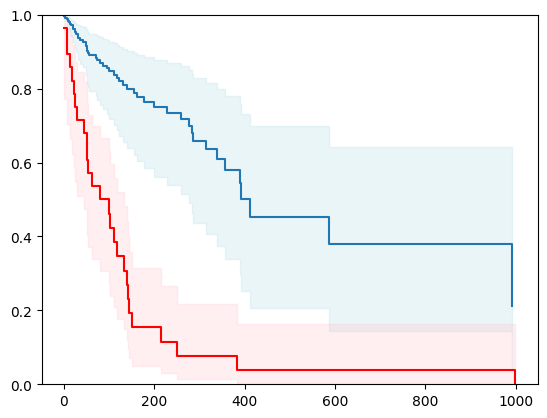
X, y = load_veterans_lung_cancer()
X = _encode_categorical_columns(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PISurvivalCustom(regr=ns.CustomRegressor(RandomForestRegressor()),
type_pi="bootstrap")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
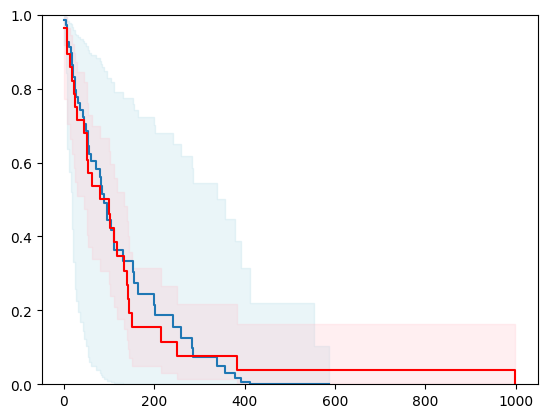
X, y = load_veterans_lung_cancer()
X = _encode_categorical_columns(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PIComponentwiseGenGradientBoostingSurvivalAnalysis(regr=ns.CustomRegressor(RidgeCV()),
type_pi="ecdf")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
100%|██████████| 100/100 [00:16<00:00, 5.95it/s]
100%|██████████| 100/100 [00:17<00:00, 5.79it/s]
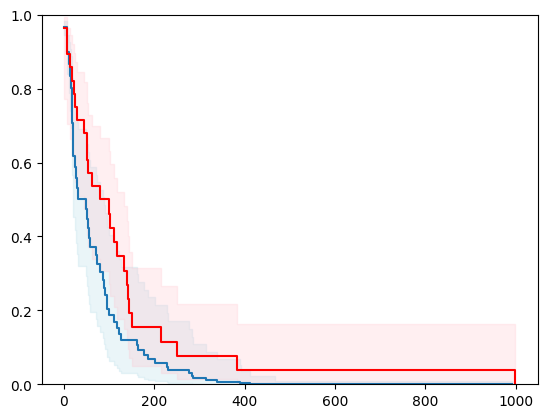
X, y = load_veterans_lung_cancer()
X = _encode_categorical_columns(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PISurvivalCustom(regr=ns.CustomRegressor(RandomForestRegressor()),
type_pi="ecdf")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
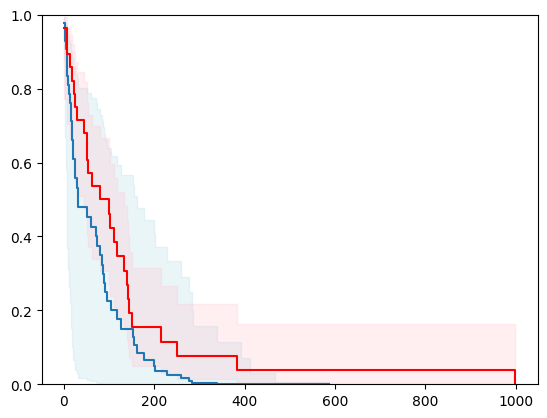
X, y = load_veterans_lung_cancer()
X = _encode_categorical_columns(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PIComponentwiseGenGradientBoostingSurvivalAnalysis(regr=ns.CustomRegressor(RidgeCV()),
type_pi="kde")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
100%|██████████| 100/100 [00:27<00:00, 3.69it/s]
100%|██████████| 100/100 [00:16<00:00, 6.04it/s]
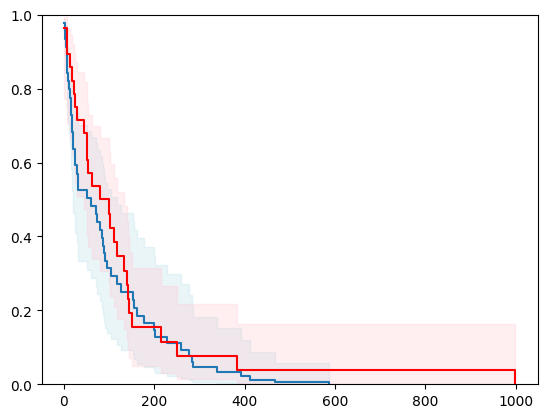
2 - 2 with Bayesian Inference
def encode_categorical_columns(df, categorical_columns=None):
"""
Automatically identifies categorical columns and applies one-hot encoding.
Parameters:
- df (pd.DataFrame): The input DataFrame with mixed continuous and categorical variables.
- categorical_columns (list): Optional list of column names to treat as categorical.
Returns:
- pd.DataFrame: A new DataFrame with one-hot encoded categorical columns.
"""
# Automatically identify categorical columns if not provided
if categorical_columns is None:
categorical_columns = df.select_dtypes(include=['object', 'category']).columns.tolist()
# Apply one-hot encoding to the identified categorical columns
df_encoded = pd.get_dummies(df, columns=categorical_columns)
# Convert boolean columns to integer (0 and 1)
bool_columns = df_encoded.select_dtypes(include=['bool']).columns.tolist()
df_encoded[bool_columns] = df_encoded[bool_columns].astype(int)
return df_encoded
X, y = load_veterans_lung_cancer()
X = encode_categorical_columns(X)
X_train, X_test, y_train, y_test = train_test_split(X, y,
test_size=0.4,
random_state=42)
print("\n\n BayesianRidge ------------------")
estimator = SurvivalCustom(regr=ns.CustomRegressor(BayesianRidge()))
estimator2 = SurvivalCustom(regr=ns.CustomRegressor(GaussianProcessRegressor()))
estimator3 = SurvivalCustom(regr=ns.CustomRegressor(ARDRegression()))
start = time()
estimator.fit(X_train, y_train)
print("Time to fit BayesianRidge: ", time() - start)
start = time()
estimator2.fit(X_train, y_train)
print("Time to fit GaussianProcessRegressor: ", time() - start)
start = time()
estimator3.fit(X_train, y_train)
print("Time to fit ARDRegression: ", time() - start)
surv_funcs = estimator.predict_survival_function(X_test.iloc[0:1,:], return_std=True)
surv_funcs2 = estimator2.predict_survival_function(X_test.iloc[0:1,:], return_std=True)
surv_funcs3 = estimator3.predict_survival_function(X_test.iloc[0:1,:], return_std=True)
BayesianRidge ------------------
Time to fit BayesianRidge: 0.17850041389465332
Time to fit GaussianProcessRegressor: 0.4104886054992676
Time to fit ARDRegression: 0.5052413940429688
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
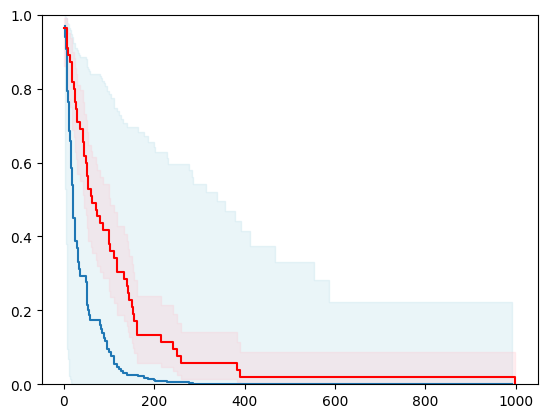
for fn in surv_funcs2.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs2.lower[0].y, surv_funcs2.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
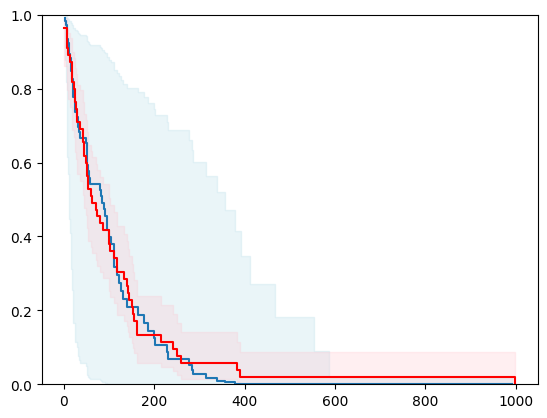
for fn in surv_funcs3.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs3.lower[0].y, surv_funcs3.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
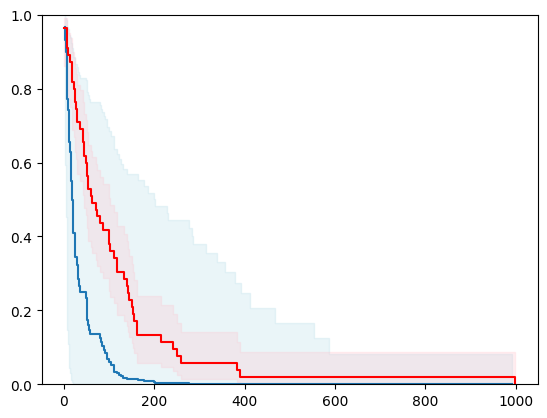
3 - using glmnet
X, y = load_veterans_lung_cancer()
X = _encode_categorical_columns(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PISurvivalCustom(regr=ns.CustomRegressor(glmnet.GLMNet(lambdau=1000)),
type_pi="bootstrap")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
estimator = PIComponentwiseGenGradientBoostingSurvivalAnalysis(regr=ns.CustomRegressor(glmnet.GLMNet(lambdau=1000)),
type_pi="kde")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
100%|██████████| 100/100 [00:36<00:00, 2.71it/s]
100%|██████████| 100/100 [00:40<00:00, 2.49it/s]
4 - using pytorch
import torch
import torch.nn as nn
import numpy as np
from sklearn.base import BaseEstimator, RegressorMixin
class MLPRegressorTorch(BaseEstimator, RegressorMixin):
def __init__(self, input_size=1, hidden_sizes=(64, 32), activation=nn.ReLU,
learning_rate=0.001, max_epochs=100, batch_size=32, random_state=None):
self.input_size = input_size
self.hidden_sizes = hidden_sizes
self.activation = activation
self.learning_rate = learning_rate
self.max_epochs = max_epochs
self.batch_size = batch_size
self.random_state = random_state
if self.random_state is not None:
torch.manual_seed(self.random_state)
def _build_model(self):
layers = []
input_dim = self.input_size
for hidden_size in self.hidden_sizes:
layers.append(nn.Linear(input_dim, hidden_size))
layers.append(self.activation())
input_dim = hidden_size
layers.append(nn.Linear(input_dim, 1)) # Output layer
self.model = nn.Sequential(*layers)
def fit(self, X, y, sample_weight=None):
if sample_weight is not None:
sample_weight = torch.tensor(sample_weight, dtype=torch.float32)
X, y = self._prepare_data(X, y)
self._build_model()
criterion = nn.MSELoss()
optimizer = torch.optim.Adam(self.model.parameters(), lr=self.learning_rate)
dataset = torch.utils.data.TensorDataset(X, y)
dataloader = torch.utils.data.DataLoader(dataset, batch_size=self.batch_size, shuffle=True)
self.model.train()
for epoch in range(self.max_epochs):
for batch_X, batch_y in dataloader:
optimizer.zero_grad()
outputs = self.model(batch_X).squeeze()
loss = criterion(outputs, batch_y)
loss.backward()
optimizer.step()
return self
def predict(self, X):
X = self._prepare_data(X)
self.model.eval()
with torch.no_grad():
predictions = self.model(X).squeeze()
return predictions.numpy()
def _prepare_data(self, X, y=None):
if isinstance(X, np.ndarray):
X = torch.tensor(X, dtype=torch.float32)
if y is not None:
if isinstance(y, np.ndarray):
y = torch.tensor(y, dtype=torch.float32)
return X, y
return X
X, y = load_veterans_lung_cancer()
X = _encode_categorical_columns(X)
# Create a new structured array with Survival_in_days as float32
new_dtype = [('Status', '?'), ('Survival_in_days', '<f4')]
y_converted = np.array(y.tolist(), dtype=new_dtype)
X_train, X_test, y_train, y_test = train_test_split(X, y_converted,
test_size=0.2,
random_state=42)
# Convert X_train and X_test to float32
X_train = X_train.astype(np.float32)
X_test = X_test.astype(np.float32)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PISurvivalCustom(regr=MLPRegressorTorch(input_size=X_train.shape[1]+1,
hidden_sizes=(20, 20, 20),
max_epochs=200,
random_state=42),
type_pi="ecdf")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
X, y = load_whas500()
X = _encode_categorical_columns(X)
# Create a new structured array with Survival_in_days as float32
new_dtype = [('Status', '?'), ('Survival_in_days', '<f4')]
y_converted = np.array(y.tolist(), dtype=new_dtype)
X_train, X_test, y_train, y_test = train_test_split(X, y_converted,
test_size=0.2,
random_state=42)
# Convert X_train and X_test to float32
X_train = X_train.astype(np.float32)
X_test = X_test.astype(np.float32)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PISurvivalCustom(regr=MLPRegressorTorch(input_size=X_train.shape[1]+1,
hidden_sizes=(20, 20, 20),
max_epochs=200,
random_state=42),
type_pi="ecdf")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
5 - Using keras (through scikeras)
import keras
import keras.models
from scikeras.wrappers import KerasRegressor
def get_reg(meta, hidden_layer_sizes, dropout):
n_features_in_ = meta["n_features_in_"]
model = keras.models.Sequential()
model.add(keras.layers.Input(shape=(n_features_in_,)))
for hidden_layer_size in hidden_layer_sizes:
model.add(keras.layers.Dense(hidden_layer_size, activation="relu"))
model.add(keras.layers.Dropout(dropout))
model.add(keras.layers.Dense(1))
return model
reg = KerasRegressor(
model=get_reg,
loss="mse",
metrics=[keras.metrics.R2Score],
hidden_layer_sizes=(20, 20, 20),
dropout=0.1,
verbose=0,
random_state=123
)
X, y = load_veterans_lung_cancer()
X = _encode_categorical_columns(X)
# Create a new structured array with Survival_in_days as float32
new_dtype = [('Status', '?'), ('Survival_in_days', '<f4')]
y_converted = np.array(y.tolist(), dtype=new_dtype)
X_train, X_test, y_train, y_test = train_test_split(X, y_converted,
test_size=0.2,
random_state=4)
X_train = X_train.astype(np.float32)
X_test = X_test.astype(np.float32)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PISurvivalCustom(regr=reg,
type_pi="kde")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()

X, y = load_whas500()
X = _encode_categorical_columns(X)
# Create a new structured array with Survival_in_days as float32
new_dtype = [('Status', '?'), ('Survival_in_days', '<f4')]
y_converted = np.array(y.tolist(), dtype=new_dtype)
X_train, X_test, y_train, y_test = train_test_split(X, y_converted,
test_size=0.2,
random_state=4)
X_train = X_train.astype(np.float32)
X_test = X_test.astype(np.float32)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PISurvivalCustom(regr=reg,
type_pi="kde")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
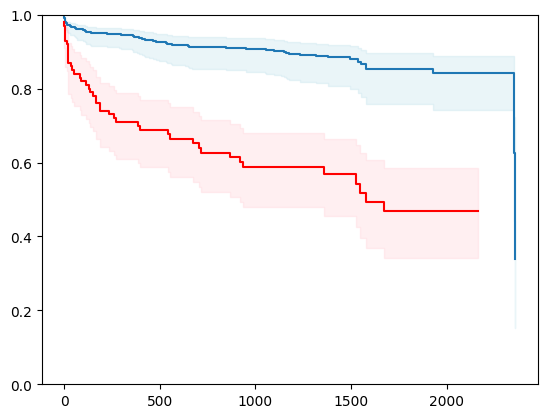
6 - using xgboost
import xgboost as xgb
X, y = load_veterans_lung_cancer()
X = _encode_categorical_columns(X)
# Create a new structured array with Survival_in_days as float32
new_dtype = [('Status', '?'), ('Survival_in_days', '<f4')]
y_converted = np.array(y.tolist(), dtype=new_dtype)
X_train, X_test, y_train, y_test = train_test_split(X, y_converted,
test_size=0.2,
random_state=4)
X_train = X_train.astype(np.float32)
X_test = X_test.astype(np.float32)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PISurvivalCustom(regr=xgb.XGBRegressor(),
type_pi="kde")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
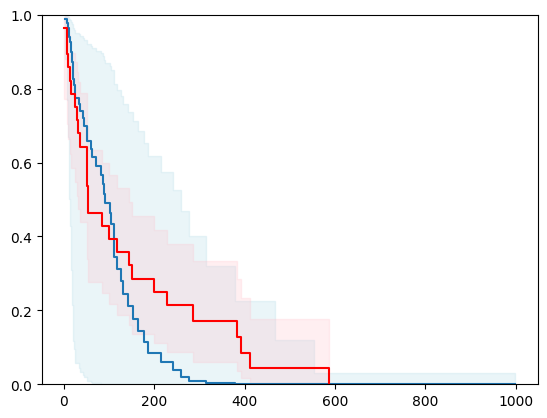
X, y = load_whas500()
X = _encode_categorical_columns(X)
# Create a new structured array with Survival_in_days as float32
new_dtype = [('Status', '?'), ('Survival_in_days', '<f4')]
y_converted = np.array(y.tolist(), dtype=new_dtype)
X_train, X_test, y_train, y_test = train_test_split(X, y_converted,
test_size=0.2,
random_state=4)
X_train = X_train.astype(np.float32)
X_test = X_test.astype(np.float32)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PISurvivalCustom(regr=xgb.XGBRegressor(),
type_pi="kde")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
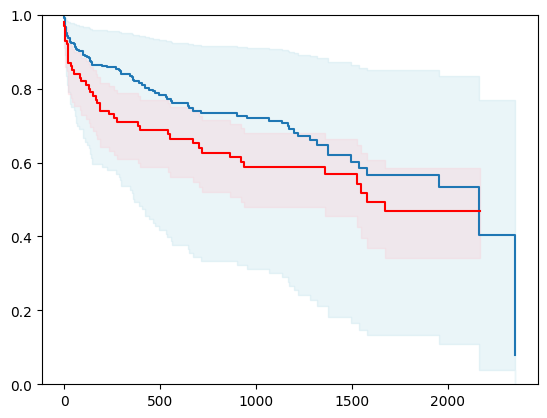
7 - using lightgbm
import lightgbm as lgb
# Create a new structured array with Survival_in_days as float32
new_dtype = [('Status', '?'), ('Survival_in_days', '<f4')]
y_converted = np.array(y.tolist(), dtype=new_dtype)
X_train, X_test, y_train, y_test = train_test_split(X, y_converted,
test_size=0.2,
random_state=4)
X_train = X_train.astype(np.float32)
X_test = X_test.astype(np.float32)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PISurvivalCustom(regr=lgb.LGBMRegressor(verbose=-1,
random_state=42),
type_pi="kde")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
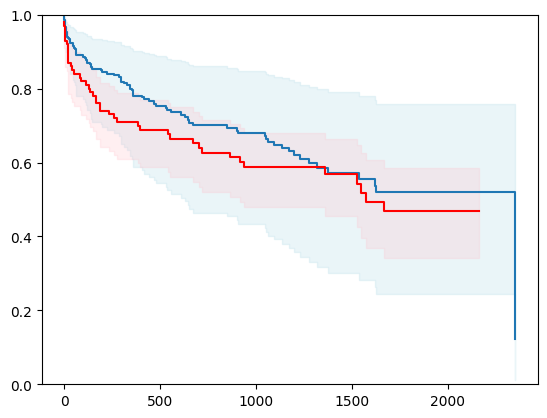
X, y = load_whas500()
X = _encode_categorical_columns(X)
# Create a new structured array with Survival_in_days as float32
new_dtype = [('Status', '?'), ('Survival_in_days', '<f4')]
y_converted = np.array(y.tolist(), dtype=new_dtype)
X_train, X_test, y_train, y_test = train_test_split(X, y_converted,
test_size=0.2,
random_state=4)
X_train = X_train.astype(np.float32)
X_test = X_test.astype(np.float32)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PISurvivalCustom(regr=lgb.LGBMRegressor(verbose=-1,
random_state=42),
type_pi="kde")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
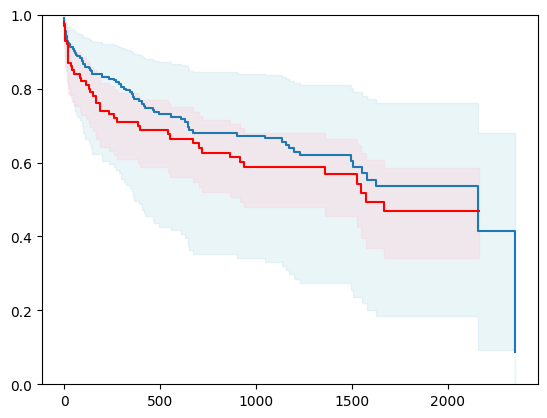
8 - using Generic Boosting (mlsauce)
import mlsauce as ms
regr_ridge = ms.GenericBoostingRegressor(ms.RidgeRegressor(reg_lambda=1e3),
verbose=0)
# Create a new structured array with Survival_in_days as float32
new_dtype = [('Status', '?'), ('Survival_in_days', '<f4')]
y_converted = np.array(y.tolist(), dtype=new_dtype)
X_train, X_test, y_train, y_test = train_test_split(X, y_converted,
test_size=0.2,
random_state=4)
X_train = X_train.astype(np.float32)
X_test = X_test.astype(np.float32)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PISurvivalCustom(regr=regr_ridge,
type_pi="kde")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
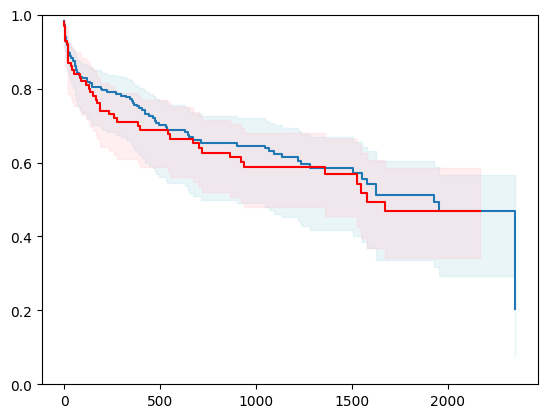
X, y = load_whas500()
X = _encode_categorical_columns(X)
# Create a new structured array with Survival_in_days as float32
new_dtype = [('Status', '?'), ('Survival_in_days', '<f4')]
y_converted = np.array(y.tolist(), dtype=new_dtype)
X_train, X_test, y_train, y_test = train_test_split(X, y_converted,
test_size=0.2,
random_state=4)
X_train = X_train.astype(np.float32)
X_test = X_test.astype(np.float32)
event_time = [y[1] for y in y_test]
event_status = [y[0] for y in y_test]
km = kaplan_meier_estimator(event_status, event_time,
conf_type="log-log")
estimator = PISurvivalCustom(regr=regr_ridge,
type_pi="kde")
estimator.fit(X_train, y_train)
surv_funcs = estimator.predict_survival_function(X_test.iloc[:1])
for fn in surv_funcs.mean:
plt.step(fn.x, fn(fn.x), where="post")
plt.fill_between(fn.x, surv_funcs.lower[0].y, surv_funcs.upper[0].y, alpha=0.25, color="lightblue", step="post")
plt.step(km[0], km[1], where="post", color="red", label="Kaplan-Meier")
plt.fill_between(km[0], km[2][0], km[2][1], alpha=0.25, color="pink", step="post")
plt.ylim(0, 1)
plt.show()
For attribution, please cite this work as:
T. Moudiki (2024-12-15). survivalist: Probabilistic model-agnostic survival analysis using scikit-learn, glmnet, xgboost, lightgbm, pytorch, keras, nnetsauce and mlsauce. Retrieved from https://thierrymoudiki.github.io/blog/2024/12/15/python/agnostic-survival-analysis
BibTeX citation (remove empty spaces)
@misc{ tmoudiki20241215,
author = { T. Moudiki },
title = { survivalist: Probabilistic model-agnostic survival analysis using scikit-learn, glmnet, xgboost, lightgbm, pytorch, keras, nnetsauce and mlsauce },
url = { https://thierrymoudiki.github.io/blog/2024/12/15/python/agnostic-survival-analysis },
year = { 2024 } }
Previous publications
- Understanding Boosted Configuration Networks (combined neural networks and boosting): An Intuitive Guide Through Their Hyperparameters Feb 16, 2026
- R version of Python package survivalist, for model-agnostic survival analysis Feb 9, 2026
- Presenting Lightweight Transfer Learning for Financial Forecasting (Risk 2026) Feb 4, 2026
- Option pricing using time series models as market price of risk Feb 1, 2026
- Enhancing Time Series Forecasting (ahead::ridge2f) with Attention-Based Context Vectors (ahead::contextridge2f) Jan 31, 2026
- Overfitting and scaling (on GPU T4) tests on nnetsauce.CustomRegressor Jan 29, 2026
- Beyond Cross-validation: Hyperparameter Optimization via Generalization Gap Modeling Jan 25, 2026
- GPopt for Machine Learning (hyperparameters' tuning) Jan 21, 2026
- rtopy: an R to Python bridge -- novelties Jan 8, 2026
- Python examples for 'Beyond Nelson-Siegel and splines: A model- agnostic Machine Learning framework for discount curve calibration, interpolation and extrapolation' Jan 3, 2026
- Forecasting benchmark: Dynrmf (a new serious competitor in town) vs Theta Method on M-Competitions and Tourism competitition Jan 1, 2026
- Finally figured out a way to port python packages to R using uv and reticulate: example with nnetsauce Dec 17, 2025
- Overfitting Random Fourier Features: Universal Approximation Property Dec 13, 2025
- Counterfactual Scenario Analysis with ahead::ridge2f Dec 11, 2025
- Zero-Shot Probabilistic Time Series Forecasting with TabPFN 2.5 and nnetsauce Dec 10, 2025
- ARIMA Pricing: Semi-Parametric Market price of risk for Risk-Neutral Pricing (code + preprint) Dec 7, 2025
- Analyzing Paper Reviews with LLMs: I Used ChatGPT, DeepSeek, Qwen, Mistral, Gemini, and Claude (and you should too + publish the analysis) Dec 3, 2025
- tisthemachinelearner: New Workflow with uv for R Integration of scikit-learn Dec 1, 2025
- (ICYMI) RPweave: Unified R + Python + LaTeX System using uv Nov 21, 2025
- unifiedml: A Unified Machine Learning Interface for R, is now on CRAN + Discussion about AI replacing humans Nov 16, 2025
- Context-aware Theta forecasting Method: Extending Classical Time Series Forecasting with Machine Learning Nov 13, 2025
- unifiedml in R: A Unified Machine Learning Interface Nov 5, 2025
- Deterministic Shift Adjustment in Arbitrage-Free Pricing (historical to risk-neutral short rates) Oct 28, 2025
- New instantaneous short rates models with their deterministic shift adjustment, for historical and risk-neutral simulation Oct 27, 2025
- RPweave: Unified R + Python + LaTeX System using uv Oct 19, 2025
- GAN-like Synthetic Data Generation Examples (on univariate, multivariate distributions, digits recognition, Fashion-MNIST, stock returns, and Olivetti faces) with DistroSimulator Oct 19, 2025
- Part2 of More data (> 150 files) on T. Moudiki's situation: a riddle/puzzle (including R, Python, bash interfaces to the game -- but everyone can play) Oct 16, 2025
- More data (> 150 files) on T. Moudiki's situation: a riddle/puzzle (including R, Python, bash interfaces to the game -- but everyone can play) Oct 12, 2025
- R port of llama2.c Oct 9, 2025
- Native uncertainty quantification for time series with NGBoost Oct 8, 2025
- NGBoost (Natural Gradient Boosting) for Regression, Classification, Time Series forecasting and Reserving Oct 6, 2025
- Real-time pricing with a pretrained probabilistic stock return model Oct 1, 2025
- Combining any model with GARCH(1,1) for probabilistic stock forecasting Sep 23, 2025
- Generating Synthetic Data with R-vine Copulas using esgtoolkit in R Sep 21, 2025
- Reimagining Equity Solvency Capital Requirement Approximation (one of my Master's Thesis subjects): From Bilinear Interpolation to Probabilistic Machine Learning Sep 16, 2025
- Transfer Learning using ahead::ridge2f on synthetic stocks returns Pt.2: synthetic data generation Sep 9, 2025
- Transfer Learning using ahead::ridge2f on synthetic stocks returns Sep 8, 2025
- I'm supposed to present 'Conformal Predictive Simulations for Univariate Time Series' at COPA CONFERENCE 2025 in London... Sep 4, 2025
- external regressors in ahead::dynrmf's interface for Machine learning forecasting Sep 1, 2025
- Another interesting decision, now for 'Beyond Nelson-Siegel and splines: A model-agnostic Machine Learning framework for discount curve calibration, interpolation and extrapolation' Aug 20, 2025
- Boosting any randomized based learner for regression, classification and univariate/multivariate time series forcasting Jul 26, 2025
- New nnetsauce version with CustomBackPropRegressor (CustomRegressor with Backpropagation) and ElasticNet2Regressor (Ridge2 with ElasticNet regularization) Jul 15, 2025
- mlsauce (home to a model-agnostic gradient boosting algorithm) can now be installed from PyPI. Jul 10, 2025
- A user-friendly graphical interface to techtonique dot net's API (will eventually contain graphics). Jul 8, 2025
- Calling =TECHTO_MLCLASSIFICATION for Machine Learning supervised CLASSIFICATION in Excel is just a matter of copying and pasting Jul 7, 2025
- Calling =TECHTO_MLREGRESSION for Machine Learning supervised regression in Excel is just a matter of copying and pasting Jul 6, 2025
- Calling =TECHTO_RESERVING and =TECHTO_MLRESERVING for claims triangle reserving in Excel is just a matter of copying and pasting Jul 5, 2025
- Calling =TECHTO_SURVIVAL for Survival Analysis in Excel is just a matter of copying and pasting Jul 4, 2025
- Calling =TECHTO_SIMULATION for Stochastic Simulation in Excel is just a matter of copying and pasting Jul 3, 2025
- Calling =TECHTO_FORECAST for forecasting in Excel is just a matter of copying and pasting Jul 2, 2025
- Random Vector Functional Link (RVFL) artificial neural network with 2 regularization parameters successfully used for forecasting/synthetic simulation in professional settings: Extensions (including Bayesian) Jul 1, 2025
- R version of 'Backpropagating quasi-randomized neural networks' Jun 24, 2025
- Backpropagating quasi-randomized neural networks Jun 23, 2025
- Beyond ARMA-GARCH: leveraging any statistical model for volatility forecasting Jun 21, 2025
- Stacked generalization (Machine Learning model stacking) + conformal prediction for forecasting with ahead::mlf Jun 18, 2025
- An Overfitting dilemma: XGBoost Default Hyperparameters vs GenericBooster + LinearRegression Default Hyperparameters Jun 14, 2025
- Programming language-agnostic reserving using RidgeCV, LightGBM, XGBoost, and ExtraTrees Machine Learning models Jun 13, 2025
- Exceptionally, and on a more personal note (otherwise I may get buried alive)... Jun 10, 2025
- Free R, Python and SQL editors in techtonique dot net Jun 9, 2025
- Beyond Nelson-Siegel and splines: A model-agnostic Machine Learning framework for discount curve calibration, interpolation and extrapolation Jun 7, 2025
- scikit-learn, glmnet, xgboost, lightgbm, pytorch, keras, nnetsauce in probabilistic Machine Learning (for longitudinal data) Reserving (work in progress) Jun 6, 2025
- R version of Probabilistic Machine Learning (for longitudinal data) Reserving (work in progress) Jun 5, 2025
- Probabilistic Machine Learning (for longitudinal data) Reserving (work in progress) Jun 4, 2025
- Python version of Beyond ARMA-GARCH: leveraging model-agnostic Quasi-Randomized networks and conformal prediction for nonparametric probabilistic stock forecasting (ML-ARCH) Jun 3, 2025
- Beyond ARMA-GARCH: leveraging model-agnostic Machine Learning and conformal prediction for nonparametric probabilistic stock forecasting (ML-ARCH) Jun 2, 2025
- Permutations and SHAPley values for feature importance in techtonique dot net's API (with R + Python + the command line) Jun 1, 2025
- Which patient is going to survive longer? Another guide to using techtonique dot net's API (with R + Python + the command line) for survival analysis May 31, 2025
- A Guide to Using techtonique.net's API and rush for simulating and plotting Stochastic Scenarios May 30, 2025
- Simulating Stochastic Scenarios with Diffusion Models: A Guide to Using techtonique.net's API for the purpose May 29, 2025
- Will my apartment in 5th avenue be overpriced or not? Harnessing the power of www.techtonique.net (+ xgboost, lightgbm, catboost) to find out May 28, 2025
- How long must I wait until something happens: A Comprehensive Guide to Survival Analysis via an API May 27, 2025
- Harnessing the Power of techtonique.net: A Comprehensive Guide to Machine Learning Classification via an API May 26, 2025
- Quantile regression with any regressor -- Examples with RandomForestRegressor, RidgeCV, KNeighborsRegressor May 20, 2025
- Survival stacking: survival analysis translated as supervised classification in R and Python May 5, 2025
- 'Bayesian' optimization of hyperparameters in a R machine learning model using the bayesianrvfl package Apr 25, 2025
- A lightweight interface to scikit-learn in R: Bayesian and Conformal prediction Apr 21, 2025
- A lightweight interface to scikit-learn in R Pt.2: probabilistic time series forecasting in conjunction with ahead::dynrmf Apr 20, 2025
- Extending the Theta forecasting method to GLMs, GAMs, GLMBOOST and attention: benchmarking on Tourism, M1, M3 and M4 competition data sets (28000 series) Apr 14, 2025
- Extending the Theta forecasting method to GLMs and attention Apr 8, 2025
- Nonlinear conformalized Generalized Linear Models (GLMs) with R package 'rvfl' (and other models) Mar 31, 2025
- Probabilistic Time Series Forecasting (predictive simulations) in Microsoft Excel using Python, xlwings lite and www.techtonique.net Mar 28, 2025
- Conformalize (improved prediction intervals and simulations) any R Machine Learning model with misc::conformalize Mar 25, 2025
- My poster for the 18th FINANCIAL RISKS INTERNATIONAL FORUM by Institut Louis Bachelier/Fondation du Risque/Europlace Institute of Finance Mar 19, 2025
- Interpretable probabilistic kernel ridge regression using Matérn 3/2 kernels Mar 16, 2025
- (News from) Probabilistic Forecasting of univariate and multivariate Time Series using Quasi-Randomized Neural Networks (Ridge2) and Conformal Prediction Mar 9, 2025
- Word-Online: re-creating Karpathy's char-RNN (with supervised linear online learning of word embeddings) for text completion Mar 8, 2025
- CRAN-like repository for most recent releases of Techtonique's R packages Mar 2, 2025
- Presenting 'Online Probabilistic Estimation of Carbon Beta and Carbon Shapley Values for Financial and Climate Risk' at Institut Louis Bachelier Feb 27, 2025
- Web app with DeepSeek R1 and Hugging Face API for chatting Feb 23, 2025
- tisthemachinelearner: A Lightweight interface to scikit-learn with 2 classes, Classifier and Regressor (in Python and R) Feb 17, 2025
- R version of survivalist: Probabilistic model-agnostic survival analysis using scikit-learn, xgboost, lightgbm (and conformal prediction) Feb 12, 2025
- Model-agnostic global Survival Prediction of Patients with Myeloid Leukemia in QRT/Gustave Roussy Challenge (challengedata.ens.fr): Python's survivalist Quickstart Feb 10, 2025
- A simple test of the martingale hypothesis in esgtoolkit Feb 3, 2025
- Command Line Interface (CLI) for techtonique.net's API Jan 31, 2025
- Gradient-Boosting and Boostrap aggregating anything (alert: high performance): Part5, easier install and Rust backend Jan 27, 2025
- Just got a paper on conformal prediction REJECTED by International Journal of Forecasting despite evidence on 30,000 time series (and more). What's going on? Part2: 1311 time series from the Tourism competition Jan 20, 2025
- Techtonique is out! (with a tutorial in various programming languages and formats) Jan 14, 2025
- Univariate and Multivariate Probabilistic Forecasting with nnetsauce and TabPFN Jan 14, 2025
- Just got a paper on conformal prediction REJECTED by International Journal of Forecasting despite evidence on 30,000 time series (and more). What's going on? Jan 5, 2025
- Python and Interactive dashboard version of Stock price forecasting with Deep Learning: throwing power at the problem (and why it won't make you rich) Dec 31, 2024
- Stock price forecasting with Deep Learning: throwing power at the problem (and why it won't make you rich) Dec 29, 2024
- No-code Machine Learning Cross-validation and Interpretability in techtonique.net Dec 23, 2024
- survivalist: Probabilistic model-agnostic survival analysis using scikit-learn, glmnet, xgboost, lightgbm, pytorch, keras, nnetsauce and mlsauce Dec 15, 2024
- Model-agnostic 'Bayesian' optimization (for hyperparameter tuning) using conformalized surrogates in GPopt Dec 9, 2024
- You can beat Forecasting LLMs (Large Language Models a.k.a foundation models) with nnetsauce.MTS Pt.2: Generic Gradient Boosting Dec 1, 2024
- You can beat Forecasting LLMs (Large Language Models a.k.a foundation models) with nnetsauce.MTS Nov 24, 2024
- Unified interface and conformal prediction (calibrated prediction intervals) for R package forecast (and 'affiliates') Nov 23, 2024
- GLMNet in Python: Generalized Linear Models Nov 18, 2024
- Gradient-Boosting anything (alert: high performance): Part4, Time series forecasting Nov 10, 2024
- Predictive scenarios simulation in R, Python and Excel using Techtonique API Nov 3, 2024
- Chat with your tabular data in www.techtonique.net Oct 30, 2024
- Gradient-Boosting anything (alert: high performance): Part3, Histogram-based boosting Oct 28, 2024
- R editor and SQL console (in addition to Python editors) in www.techtonique.net Oct 21, 2024
- R and Python consoles + JupyterLite in www.techtonique.net Oct 15, 2024
- Gradient-Boosting anything (alert: high performance): Part2, R version Oct 14, 2024
- Gradient-Boosting anything (alert: high performance) Oct 6, 2024
- Benchmarking 30 statistical/Machine Learning models on the VN1 Forecasting -- Accuracy challenge Oct 4, 2024
- Automated random variable distribution inference using Kullback-Leibler divergence and simulating best-fitting distribution Oct 2, 2024
- Forecasting in Excel using Techtonique's Machine Learning APIs under the hood Sep 30, 2024
- Techtonique web app for data-driven decisions using Mathematics, Statistics, Machine Learning, and Data Visualization Sep 25, 2024
- Parallel for loops (Map or Reduce) + New versions of nnetsauce and ahead Sep 16, 2024
- Adaptive (online/streaming) learning with uncertainty quantification using Polyak averaging in learningmachine Sep 10, 2024
- New versions of nnetsauce and ahead Sep 9, 2024
- Prediction sets and prediction intervals for conformalized Auto XGBoost, Auto LightGBM, Auto CatBoost, Auto GradientBoosting Sep 2, 2024
- Quick/automated R package development workflow (assuming you're using macOS or Linux) Part2 Aug 30, 2024
- R package development workflow (assuming you're using macOS or Linux) Aug 27, 2024
- A new method for deriving a nonparametric confidence interval for the mean Aug 26, 2024
- Conformalized adaptive (online/streaming) learning using learningmachine in Python and R Aug 19, 2024
- Bayesian (nonlinear) adaptive learning Aug 12, 2024
- Auto XGBoost, Auto LightGBM, Auto CatBoost, Auto GradientBoosting Aug 5, 2024
- Copulas for uncertainty quantification in time series forecasting Jul 28, 2024
- Forecasting uncertainty: sequential split conformal prediction + Block bootstrap (web app) Jul 22, 2024
- learningmachine for Python (new version) Jul 15, 2024
- learningmachine v2.0.0: Machine Learning with explanations and uncertainty quantification Jul 8, 2024
- My presentation at ISF 2024 conference (slides with nnetsauce probabilistic forecasting news) Jul 3, 2024
- 10 uncertainty quantification methods in nnetsauce forecasting Jul 1, 2024
- Forecasting with XGBoost embedded in Quasi-Randomized Neural Networks Jun 24, 2024
- Forecasting Monthly Airline Passenger Numbers with Quasi-Randomized Neural Networks Jun 17, 2024
- Automated hyperparameter tuning using any conformalized surrogate Jun 9, 2024
- Recognizing handwritten digits with Ridge2Classifier Jun 3, 2024
- Forecasting the Economy May 27, 2024
- A detailed introduction to Deep Quasi-Randomized 'neural' networks May 19, 2024
- Probability of receiving a loan; using learningmachine May 12, 2024
- mlsauce's `v0.18.2`: various examples and benchmarks with dimension reduction May 6, 2024
- mlsauce's `v0.17.0`: boosting with Elastic Net, polynomials and heterogeneity in explanatory variables Apr 29, 2024
- mlsauce's `v0.13.0`: taking into account inputs heterogeneity through clustering Apr 21, 2024
- mlsauce's `v0.12.0`: prediction intervals for LSBoostRegressor Apr 15, 2024
- Conformalized predictive simulations for univariate time series on more than 250 data sets Apr 7, 2024
- learningmachine v1.1.2: for Python Apr 1, 2024
- learningmachine v1.0.0: prediction intervals around the probability of the event 'a tumor being malignant' Mar 25, 2024
- Bayesian inference and conformal prediction (prediction intervals) in nnetsauce v0.18.1 Mar 18, 2024
- Multiple examples of Machine Learning forecasting with ahead Mar 11, 2024
- rtopy (v0.1.1): calling R functions in Python Mar 4, 2024
- ahead forecasting (v0.10.0): fast time series model calibration and Python plots Feb 26, 2024
- A plethora of datasets at your fingertips Part3: how many times do couples cheat on each other? Feb 19, 2024
- nnetsauce's introduction as of 2024-02-11 (new version 0.17.0) Feb 11, 2024
- Tuning Machine Learning models with GPopt's new version Part 2 Feb 5, 2024
- Tuning Machine Learning models with GPopt's new version Jan 29, 2024
- Subsampling continuous and discrete response variables Jan 22, 2024
- DeepMTS, a Deep Learning Model for Multivariate Time Series Jan 15, 2024
- A classifier that's very accurate (and deep) Pt.2: there are > 90 classifiers in nnetsauce Jan 8, 2024
- learningmachine: prediction intervals for conformalized Kernel ridge regression and Random Forest Jan 1, 2024
- A plethora of datasets at your fingertips Part2: how many times do couples cheat on each other? Descriptive analytics, interpretability and prediction intervals using conformal prediction Dec 25, 2023
- Diffusion models in Python with esgtoolkit (Part2) Dec 18, 2023
- Diffusion models in Python with esgtoolkit Dec 11, 2023
- Julia packaging at the command line Dec 4, 2023
- Quasi-randomized nnetworks in Julia, Python and R Nov 27, 2023
- A plethora of datasets at your fingertips Nov 20, 2023
- A classifier that's very accurate (and deep) Nov 12, 2023
- mlsauce version 0.8.10: Statistical/Machine Learning with Python and R Nov 5, 2023
- AutoML in nnetsauce (randomized and quasi-randomized nnetworks) Pt.2: multivariate time series forecasting Oct 29, 2023
- AutoML in nnetsauce (randomized and quasi-randomized nnetworks) Oct 22, 2023
- Version v0.14.0 of nnetsauce for R and Python Oct 16, 2023
- A diffusion model: G2++ Oct 9, 2023
- Diffusion models in ESGtoolkit + announcements Oct 2, 2023
- An infinity of time series forecasting models in nnetsauce (Part 2 with uncertainty quantification) Sep 25, 2023
- (News from) forecasting in Python with ahead (progress bars and plots) Sep 18, 2023
- Forecasting in Python with ahead Sep 11, 2023
- Risk-neutralize simulations Sep 4, 2023
- Comparing cross-validation results using crossval_ml and boxplots Aug 27, 2023
- Reminder Apr 30, 2023
- Did you ask ChatGPT about who you are? Apr 16, 2023
- A new version of nnetsauce (randomized and quasi-randomized 'neural' networks) Apr 2, 2023
- Simple interfaces to the forecasting API Nov 23, 2022
- A web application for forecasting in Python, R, Ruby, C#, JavaScript, PHP, Go, Rust, Java, MATLAB, etc. Nov 2, 2022
- Prediction intervals (not only) for Boosted Configuration Networks in Python Oct 5, 2022
- Boosted Configuration (neural) Networks Pt. 2 Sep 3, 2022
- Boosted Configuration (_neural_) Networks for classification Jul 21, 2022
- A Machine Learning workflow using Techtonique Jun 6, 2022
- Super Mario Bros © in the browser using PyScript May 8, 2022
- News from ESGtoolkit, ycinterextra, and nnetsauce Apr 4, 2022
- Explaining a Keras _neural_ network predictions with the-teller Mar 11, 2022
- New version of nnetsauce -- various quasi-randomized networks Feb 12, 2022
- A dashboard illustrating bivariate time series forecasting with `ahead` Jan 14, 2022
- Hundreds of Statistical/Machine Learning models for univariate time series, using ahead, ranger, xgboost, and caret Dec 20, 2021
- Forecasting with `ahead` (Python version) Dec 13, 2021
- Tuning and interpreting LSBoost Nov 15, 2021
- Time series cross-validation using `crossvalidation` (Part 2) Nov 7, 2021
- Fast and scalable forecasting with ahead::ridge2f Oct 31, 2021
- Automatic Forecasting with `ahead::dynrmf` and Ridge regression Oct 22, 2021
- Forecasting with `ahead` Oct 15, 2021
- Classification using linear regression Sep 26, 2021
- `crossvalidation` and random search for calibrating support vector machines Aug 6, 2021
- parallel grid search cross-validation using `crossvalidation` Jul 31, 2021
- `crossvalidation` on R-universe, plus a classification example Jul 23, 2021
- Documentation and source code for GPopt, a package for Bayesian optimization Jul 2, 2021
- Hyperparameters tuning with GPopt Jun 11, 2021
- A forecasting tool (API) with examples in curl, R, Python May 28, 2021
- Bayesian Optimization with GPopt Part 2 (save and resume) Apr 30, 2021
- Bayesian Optimization with GPopt Apr 16, 2021
- Compatibility of nnetsauce and mlsauce with scikit-learn Mar 26, 2021
- Explaining xgboost predictions with the teller Mar 12, 2021
- An infinity of time series models in nnetsauce Mar 6, 2021
- New activation functions in mlsauce's LSBoost Feb 12, 2021
- 2020 recap, Gradient Boosting, Generalized Linear Models, AdaOpt with nnetsauce and mlsauce Dec 29, 2020
- A deeper learning architecture in nnetsauce Dec 18, 2020
- Classify penguins with nnetsauce's MultitaskClassifier Dec 11, 2020
- Bayesian forecasting for uni/multivariate time series Dec 4, 2020
- Generalized nonlinear models in nnetsauce Nov 28, 2020
- Boosting nonlinear penalized least squares Nov 21, 2020
- Statistical/Machine Learning explainability using Kernel Ridge Regression surrogates Nov 6, 2020
- NEWS Oct 30, 2020
- A glimpse into my PhD journey Oct 23, 2020
- Submitting R package to CRAN Oct 16, 2020
- Simulation of dependent variables in ESGtoolkit Oct 9, 2020
- Forecasting lung disease progression Oct 2, 2020
- New nnetsauce Sep 25, 2020
- Technical documentation Sep 18, 2020
- A new version of nnetsauce, and a new Techtonique website Sep 11, 2020
- Back next week, and a few announcements Sep 4, 2020
- Explainable 'AI' using Gradient Boosted randomized networks Pt2 (the Lasso) Jul 31, 2020
- LSBoost: Explainable 'AI' using Gradient Boosted randomized networks (with examples in R and Python) Jul 24, 2020
- nnetsauce version 0.5.0, randomized neural networks on GPU Jul 17, 2020
- Maximizing your tip as a waiter (Part 2) Jul 10, 2020
- New version of mlsauce, with Gradient Boosted randomized networks and stump decision trees Jul 3, 2020
- Announcements Jun 26, 2020
- Parallel AdaOpt classification Jun 19, 2020
- Comments section and other news Jun 12, 2020
- Maximizing your tip as a waiter Jun 5, 2020
- AdaOpt classification on MNIST handwritten digits (without preprocessing) May 29, 2020
- AdaOpt (a probabilistic classifier based on a mix of multivariable optimization and nearest neighbors) for R May 22, 2020
- AdaOpt May 15, 2020
- Custom errors for cross-validation using crossval::crossval_ml May 8, 2020
- Documentation+Pypi for the `teller`, a model-agnostic tool for Machine Learning explainability May 1, 2020
- Encoding your categorical variables based on the response variable and correlations Apr 24, 2020
- Linear model, xgboost and randomForest cross-validation using crossval::crossval_ml Apr 17, 2020
- Grid search cross-validation using crossval Apr 10, 2020
- Documentation for the querier, a query language for Data Frames Apr 3, 2020
- Time series cross-validation using crossval Mar 27, 2020
- On model specification, identification, degrees of freedom and regularization Mar 20, 2020
- Import data into the querier (now on Pypi), a query language for Data Frames Mar 13, 2020
- R notebooks for nnetsauce Mar 6, 2020
- Version 0.4.0 of nnetsauce, with fruits and breast cancer classification Feb 28, 2020
- Create a specific feed in your Jekyll blog Feb 21, 2020
- Git/Github for contributing to package development Feb 14, 2020
- Feedback forms for contributing Feb 7, 2020
- nnetsauce for R Jan 31, 2020
- A new version of nnetsauce (v0.3.1) Jan 24, 2020
- ESGtoolkit, a tool for Monte Carlo simulation (v0.2.0) Jan 17, 2020
- Search bar, new year 2020 Jan 10, 2020
- 2019 Recap, the nnetsauce, the teller and the querier Dec 20, 2019
- Understanding model interactions with the `teller` Dec 13, 2019
- Using the `teller` on a classifier Dec 6, 2019
- Benchmarking the querier's verbs Nov 29, 2019
- Composing the querier's verbs for data wrangling Nov 22, 2019
- Comparing and explaining model predictions with the teller Nov 15, 2019
- Tests for the significance of marginal effects in the teller Nov 8, 2019
- Introducing the teller Nov 1, 2019
- Introducing the querier Oct 25, 2019
- Prediction intervals for nnetsauce models Oct 18, 2019
- Using R in Python for statistical learning/data science Oct 11, 2019
- Model calibration with `crossval` Oct 4, 2019
- Bagging in the nnetsauce Sep 25, 2019
- Adaboost learning with nnetsauce Sep 18, 2019
- Change in blog's presentation Sep 4, 2019
- nnetsauce on Pypi Jun 5, 2019
- More nnetsauce (examples of use) May 9, 2019
- nnetsauce Mar 13, 2019
- crossval Mar 13, 2019
- test Mar 10, 2019


Comments powered by Talkyard.