I created R package learningmachine in the first place, in order to have a unified, object-oriented (using R6), interface for the machine learning algorithms I use the most on tabular data. This (work in progress) package is available on GitHub and the R-universe. There will certainly be a Python version in the future.
This post shows how to use learningmachine to compute prediction intervals for miles per gallon (mpg) car consumption using conformalized Kernel ridge regression and R package ranger’s Random Forest.
Install packages
utils::install.packages('learningmachine',
repos = c('https://techtonique.r-universe.dev',
'https://cloud.r-project.org'))
utils::install.packages("skimr")
Import dataset
data(mtcars)
Descriptive statistics
skimr::skim(mtcars)
── Data Summary ────────────────────────
Values
Name mtcars
Number of rows 32
Number of columns 11
_______________________
Column type frequency:
numeric 11
________________________
Group variables None
── Variable type: numeric ──────────────────────────────────────────────────────
skim_variable n_missing complete_rate mean sd p0 p25 p50
1 mpg 0 1 20.1 6.03 10.4 15.4 19.2
2 cyl 0 1 6.19 1.79 4 4 6
3 disp 0 1 231. 124. 71.1 121. 196.
4 hp 0 1 147. 68.6 52 96.5 123
5 drat 0 1 3.60 0.535 2.76 3.08 3.70
6 wt 0 1 3.22 0.978 1.51 2.58 3.32
7 qsec 0 1 17.8 1.79 14.5 16.9 17.7
8 vs 0 1 0.438 0.504 0 0 0
9 am 0 1 0.406 0.499 0 0 0
10 gear 0 1 3.69 0.738 3 3 4
11 carb 0 1 2.81 1.62 1 2 2
p75 p100 hist
1 22.8 33.9 ▃▇▅▁▂
2 8 8 ▆▁▃▁▇
3 326 472 ▇▃▃▃▂
4 180 335 ▇▇▆▃▁
5 3.92 4.93 ▇▃▇▅▁
6 3.61 5.42 ▃▃▇▁▂
7 18.9 22.9 ▃▇▇▂▁
8 1 1 ▇▁▁▁▆
9 1 1 ▇▁▁▁▆
10 4 5 ▇▁▆▁▂
11 4 8 ▇▂▅▁▁
Model fitting and predictions
library(learningmachine)
## Data -----------------------------------------------------------------------------
X <- as.matrix(mtcars[,-1])
y <- mtcars$mpg
# Split train/test
set.seed(123)
(index_train <- base::sample.int(n = nrow(X),
size = floor(0.7*nrow(X)),
replace = FALSE))
X_train <- X[index_train, ]
y_train <- y[index_train]
X_test <- X[-index_train, ]
y_test <- y[-index_train]
dim(X_train)
dim(X_test)
## Kernel Ridge Regressor (KRR) and Random Forest (RF) objects -----------------------------------------------------------------------------
obj_KRR <- learningmachine::KernelRidgeRegressor$new()
obj_RF <- learningmachine::RangerRegressor$new()
## Fit KRR and RF -----------------------------------------------------------------------------
t0 <- proc.time()[3]
obj_KRR$fit(X_train, y_train, lambda = 0.05)
cat("Elapsed: ", proc.time()[3] - t0, "s \n")
t0 <- proc.time()[3]
obj_RF$fit(X_train, y_train)
cat("Elapsed: ", proc.time()[3] - t0, "s \n")
## Predictions ------------------------------------------------------------
res_KRR <- obj_KRR$predict(X = X_test, level = 95,
method = "splitconformal")
res2_KRR <- obj_KRR$predict(X = X_test, level = 95,
method = "jackknifeplus")
res_RF <- obj_RF$predict(X = X_test, level = 95,
method = "splitconformal")
res2_RF <- obj_RF$predict(X = X_test, level = 95,
method = "jackknifeplus")
Elapsed: 0.005 s
Elapsed: 0.058 s
|======================================================================| 100%
|======================================================================| 100%
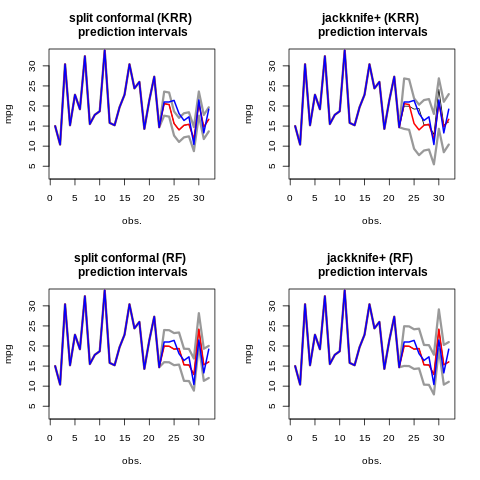
Graph
par(mfrow=c(2, 2))
plot(c(y_train, res_KRR$preds), type='l',
main="split conformal (KRR) \n prediction intervals",
xlab="obs.",
ylab="mpg",
ylim = c(3, 33))
lines(c(y_train, res_KRR$upper), col="gray60", lwd = 3)
lines(c(y_train, res_KRR$lower), col="gray60", lwd = 3)
lines(c(y_train, res_KRR$preds), col = "red", lwd = 2)
lines(c(y_train, y_test), col = "blue", lwd = 2)
plot(c(y_train, res2_KRR$preds), type='l',
main="jackknife+ (KRR) \n prediction intervals",
xlab="obs.",
ylab="mpg",
ylim = c(3, 33))
lines(c(y_train, res2_KRR$upper), col="gray60", lwd = 3)
lines(c(y_train, res2_KRR$lower), col="gray60", lwd = 3)
lines(c(y_train, res2_KRR$preds), col = "red", lwd = 2)
lines(c(y_train, y_test), col = "blue", lwd = 2)
plot(c(y_train, res_RF$preds), type='l',
main="split conformal (RF) \n prediction intervals",
xlab="obs.",
ylab="mpg",
ylim = c(3, 33))
lines(c(y_train, res_RF$upper), col="gray60", lwd = 3)
lines(c(y_train, res_RF$lower), col="gray60", lwd = 3)
lines(c(y_train, res_RF$preds), col = "red", lwd = 2)
lines(c(y_train, y_test), col = "blue", lwd = 2)
plot(c(y_train, res2_RF$preds), type='l',
main="jackknife+ (RF) \n prediction intervals",
xlab="obs.",
ylab="mpg",
ylim = c(3, 33))
lines(c(y_train, res2_RF$upper), col="gray60", lwd = 3)
lines(c(y_train, res2_RF$lower), col="gray60", lwd = 3)
lines(c(y_train, res2_RF$preds), col = "red", lwd = 2)
lines(c(y_train, y_test), col = "blue", lwd = 2)