Today, give a try to Techtonique web app, a tool designed to help you make informed, data-driven decisions using Mathematics, Statistics, Machine Learning, and Data Visualization. Here is a tutorial with audio, video, code, and slides: https://moudiki2.gumroad.com/l/nrhgb. 100 API requests are now (and forever) offered to every user every month, no matter the pricing tier.
Sometimes in Statistical/Machine Learning problems, we encounter categorical explanatory variables with high cardinality. Let’s say for example that we want to determine if a diet is good or bad, based on what a person eats. In trying to answer this question, we’d construct a response variable containing a sequence of characters good or bad, one for each person; and an explanatory variable for the model would be:
x = c("apple", "tomato", "banana", "apple", "pineapple", "bic mac",
"banana", "bic mac", "quinoa sans gluten", "pineapple",
"avocado", "avocado", "avocado", "avocado!", ...)
Some Statistical/Machine learning models only accept numerical data as input. Hence the need for a way to transform those categorical inputs into numerical vectors. One way to deal with a covariate such as x is to use one-hot encoding, as depicted below:
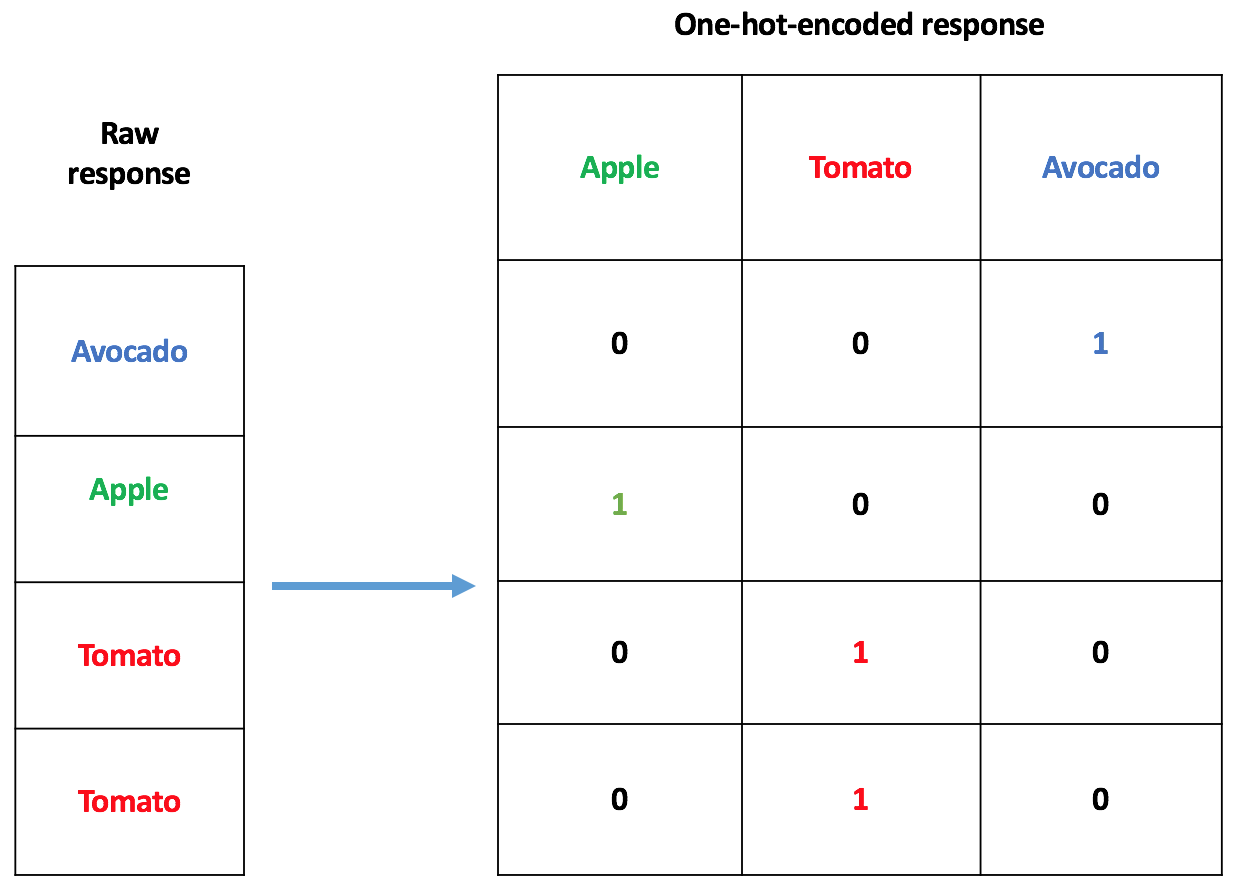
In the case of x having 100 types of fruits in it, one-hot encoding will lead to 99 explanatory variables for the model, instead of, possibly one. This means: more disk space required, more computer memory needed, and a longer training time. Apart from the one-hot encoder, there are a lot of categorical encoders out there. I wanted a relatively simple one, so I came up with the one described in this post. It’s a target-based categorical encoder, which makes use of the correlation between a randomly generated pseudo-target and the real target (a.k.a response; a sequence good or bads as seen before).
Data and packages for the demo
We’ll be using the CO2 dataset available in base R for this demo. According to its description: the CO2 data frame has 84 rows and 5 columns of data from an experiment on the cold tolerance of the grass species Echinochloa crus-gall.
# Packages required
library(randomForest)
# Dataset
Xy <- datasets::CO2
Xy$uptake <- scale(Xy$uptake) # centering and scaling the response
print(dim(Xy))
print(head(Xy))
print(tail(Xy))
Now we create a response variables and covariates, based on CO2 data:
y <- Xy$uptake
X <- Xy[, c("Plant", "Type", "Treatment" ,"conc")]
First encoder: “One-hot”
Using base R’s function model.matrix, we transform the categorical variables from CO2 to numerical variables. It’s not exactly “One-hot” as we described it previously, but a close cousin, because the covariate Plant possesses some sort of ordering (it’s “an ordered factor with levels Qn1 < Qn2 < Qn3 < … < Mc1 giving a unique identifier for each plant”):
X_onehot <- model.matrix(uptake ~ ., data=CO2)[,-1]
print(dim(X_onehot))
print(head(X_onehot))
print(tail(X_onehot))
## [1] 84 14
## Plant.L Plant.Q Plant.C Plant^4 Plant^5 Plant^6
## 1 -0.4599331 0.5018282 -0.4599331 0.3687669 -0.2616083 0.1641974
## 2 -0.4599331 0.5018282 -0.4599331 0.3687669 -0.2616083 0.1641974
## 3 -0.4599331 0.5018282 -0.4599331 0.3687669 -0.2616083 0.1641974
## 4 -0.4599331 0.5018282 -0.4599331 0.3687669 -0.2616083 0.1641974
## 5 -0.4599331 0.5018282 -0.4599331 0.3687669 -0.2616083 0.1641974
## 6 -0.4599331 0.5018282 -0.4599331 0.3687669 -0.2616083 0.1641974
## Plant^7 Plant^8 Plant^9 Plant^10 Plant^11
## 1 -0.09047913 0.04307668 -0.01721256 0.005456097 -0.001190618
## 2 -0.09047913 0.04307668 -0.01721256 0.005456097 -0.001190618
## 3 -0.09047913 0.04307668 -0.01721256 0.005456097 -0.001190618
## 4 -0.09047913 0.04307668 -0.01721256 0.005456097 -0.001190618
## 5 -0.09047913 0.04307668 -0.01721256 0.005456097 -0.001190618
## 6 -0.09047913 0.04307668 -0.01721256 0.005456097 -0.001190618
## TypeMississippi Treatmentchilled conc
## 1 0 0 95
## 2 0 0 175
## 3 0 0 250
## 4 0 0 350
## 5 0 0 500
## 6 0 0 675
## Plant.L Plant.Q Plant.C Plant^4 Plant^5 Plant^6
## 79 0.3763089 0.2281037 -0.0418121 -0.3017184 -0.4518689 -0.4627381
## 80 0.3763089 0.2281037 -0.0418121 -0.3017184 -0.4518689 -0.4627381
## 81 0.3763089 0.2281037 -0.0418121 -0.3017184 -0.4518689 -0.4627381
## 82 0.3763089 0.2281037 -0.0418121 -0.3017184 -0.4518689 -0.4627381
## 83 0.3763089 0.2281037 -0.0418121 -0.3017184 -0.4518689 -0.4627381
## 84 0.3763089 0.2281037 -0.0418121 -0.3017184 -0.4518689 -0.4627381
## Plant^7 Plant^8 Plant^9 Plant^10 Plant^11 TypeMississippi
## 79 -0.3701419 -0.2388798 -0.1236175 -0.04910487 -0.0130968 1
## 80 -0.3701419 -0.2388798 -0.1236175 -0.04910487 -0.0130968 1
## 81 -0.3701419 -0.2388798 -0.1236175 -0.04910487 -0.0130968 1
## 82 -0.3701419 -0.2388798 -0.1236175 -0.04910487 -0.0130968 1
## 83 -0.3701419 -0.2388798 -0.1236175 -0.04910487 -0.0130968 1
## 84 -0.3701419 -0.2388798 -0.1236175 -0.04910487 -0.0130968 1
## Treatmentchilled conc
## 79 1 175
## 80 1 250
## 81 1 350
## 82 1 500
## 83 1 675
## 84 1 1000
Second encoder: Target-based
Now, we present the encoder discussed in the introduction. It’s a target-based categorical encoder, which uses the correlation between a randomly generated pseudo-target and the real target.
Construction of a pseudo-target via Cholesky decomposition
Most target encoders rely directly on the response variable, which leads to a potential risk called leakage. Target encoding is indeed a form of more or less subtle overfitting. Here, in order to somehow circumvent this issue, we use Cholesky decomposition. We create a pseudo-target based on the real target uptake (centered and scaled, and stored in variable y), and specifically ask that, this pseudo-target has a fixed correlation of -0.4 (could be anything) with the response:
# reproducibility seed
set.seed(518)
# target covariance matrix
rho <- -0.4 # desired target
C <- matrix(rep(rho, 4), nrow = 2, ncol = 2)
diag(C) <- 1
# Cholesky decomposition
(C_ <- chol(C))
print(t(C_)%*%C_)
X2 <- rnorm(n)
XX <- cbind(y, X2)
# induce correlation through Cholesky decomposition
X_ <- XX %*% C_
colnames(X_) <- c("real_target", "pseudo_target")
Print the induced correlation between the randomly generated pseudo-target and the real target:
cor(y, X_[,2])
## [,1]
## [1,] -0.4008563
Now, a glimpse at X_, a matrix containing the real target and the pseudo target in columns:
print(dim(X_))
print(head(X_))
print(tail(X_))
## [1] 84 2
## real_target pseudo_target
## [1,] -1.0368659 -0.6668123
## [2,] 0.2946905 0.3894672
## [3,] 0.7015550 0.3485984
## [4,] 0.9234810 -1.2769424
## [5,] 0.7477896 -1.1996023
## [6,] 1.1084194 0.4008157
## real_target pseudo_target
## [79,] -0.8519275 -0.3455701
## [80,] -0.8611744 1.7142739
## [81,] -0.8611744 0.1521795
## [82,] -0.8611744 -0.3912856
## [83,] -0.7687052 0.9726421
## [84,] -0.6762360 0.8791499
A few checks
By repeating the procedure that we just outlined with 1000 seeds going from 1 to 1000, we obtain a distribution of achieved correlations between the real target and the pseudo target:
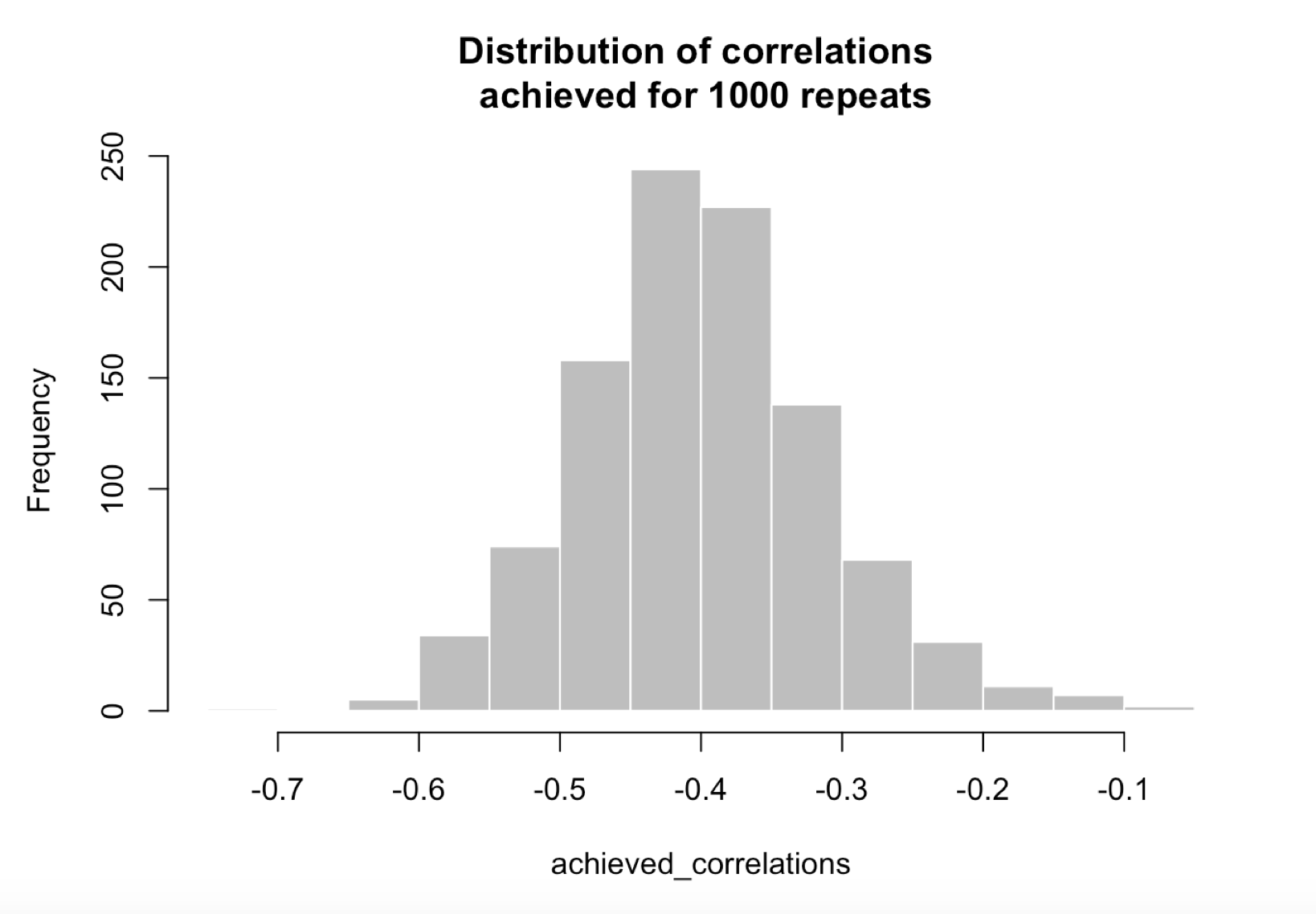
## $breaks
## [1] -0.75 -0.70 -0.65 -0.60 -0.55 -0.50 -0.45 -0.40 -0.35 -0.30 -0.25
## [12] -0.20 -0.15 -0.10 -0.05
##
## $counts
## [1] 1 0 5 34 74 158 244 227 138 68 31 11 7 2
##
## $density
## [1] 0.02 0.00 0.10 0.68 1.48 3.16 4.88 4.54 2.76 1.36 0.62 0.22 0.14 0.04
##
## $mids
## [1] -0.725 -0.675 -0.625 -0.575 -0.525 -0.475 -0.425 -0.375 -0.325 -0.275
## [11] -0.225 -0.175 -0.125 -0.075
##
## $xname
## [1] "achieved_correlations"
##
## $equidist
## [1] TRUE
##
## attr(,"class")
## [1] "histogram"
print(summary(achieved_correlations))
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## -0.70120 -0.45510 -0.40270 -0.40040 -0.34820 -0.08723
Encoding
In order to encode the factors, we use the pseudo-target y_ defined as:
y_ <- X_[ , 'pseudo_target']
Our new, numerically encoded covariates are derived by calculating sums of the pseudo-target y_ (we could think of other types of aggregations), groupped by factor level for each factor. The new matrix of covariates is named X_Cholesky:
print(dim(X_Cholesky))
print(head(X_Cholesky))
print(tail(X_Cholesky))
## [1] 84 4
## Plant Type Treatment conc
## [1,] -1.853112 -18.08574 -7.508514 95
## [2,] -1.853112 -18.08574 -7.508514 175
## [3,] -1.853112 -18.08574 -7.508514 250
## [4,] -1.853112 -18.08574 -7.508514 350
## [5,] -1.853112 -18.08574 -7.508514 500
## [6,] -1.853112 -18.08574 -7.508514 675
## Plant Type Treatment conc
## [79,] 2.628531 9.658954 -0.9182766 175
## [80,] 2.628531 9.658954 -0.9182766 250
## [81,] 2.628531 9.658954 -0.9182766 350
## [82,] 2.628531 9.658954 -0.9182766 500
## [83,] 2.628531 9.658954 -0.9182766 675
## [84,] 2.628531 9.658954 -0.9182766 1000
Notice that X_Cholesky has 4 covariates, that X_onehot had 14 covariates, and imagine a situation with a higher cardinality for each factor.
Fit a model to one-hot encoded and target based covariates
In this section, we compare both types of encoding using cross-validation with Root Mean Squared Errors (RMSE).
Datasets
# Dataset with one-hot encoded covariates
Xy1 <- data.frame(y, X_onehot)
# Dataset with pseudo-target-based encoding of covariates
Xy2 <- data.frame(y, X_Cholesky)
Comparison
Using a Random Forest here as a simple illustration without hyperparameter tuning, but tree-based models will typically handle this type of data. Not linear models, nor Neural Networks or Support Vector Machines.
Random Forests with 100 seeds, going from 1 to 100 are adjusted:
n_reps <- 100
n_train <- length(y)
`%op%` <- foreach::`%do%`
pb <- utils::txtProgressBar(min=0, max=n_reps, style = 3)
errs <- foreach::foreach(i = 1:n_reps, .combine=rbind)%op%
{
# utils::setTxtProgressBar(pb, i)
set.seed(i)
index_train <- sample.int(n_train, size = floor(0.8*n_train))
obj1 <- randomForest(y ~ ., data=Xy1[index_train, ])
obj2 <- randomForest(y ~ ., data=Xy2[index_train, ])
c(sqrt(mean((predict(obj1, newdata=as.matrix(Xy1[-index_train, -1])) - y[-index_train])^2)),
sqrt(mean((predict(obj2, newdata=as.matrix(Xy2[-index_train, -1])) - y[-index_train])^2)))
}
close(pb)
colnames(errs) <- c("one-hot", "target-based")
print(colMeans(errs))
print(apply(errs, 2, sd))
print(sapply(1:2, function (j) summary(errs[,j])))
## one-hot target-based
## 0.4121657 0.4574857
## one-hot target-based
## 0.09710344 0.07584037
## [,1] [,2]
## Min. 0.1877 0.2850
## 1st Qu. 0.3566 0.4039
## Median 0.4037 0.4464
## Mean 0.4122 0.4575
## 3rd Qu. 0.4784 0.4913
## Max. 0.6470 0.6840
There are certainly some improvements to be brought to this methodology, but the results discussed in this post already look quite encouraging to me.
Note: I am currently looking for a gig. You can hire me on Malt or send me an email: thierry dot moudiki at pm dot me. I can do descriptive statistics, data preparation, feature engineering, model calibration, training and validation, and model outputs’ interpretation. I am fluent in Python, R, SQL, Microsoft Excel, Visual Basic (among others) and French. My résumé? Here!
For attribution, please cite this work as:
T. Moudiki (2020-04-24). Encoding your categorical variables based on the response variable and correlations. Retrieved from https://thierrymoudiki.github.io/blog/2020/04/24/python/r/misc/target-encoder-correlation
BibTeX citation (remove empty spaces)
@misc{ tmoudiki20200424,
author = { T. Moudiki },
title = { Encoding your categorical variables based on the response variable and correlations },
url = { https://thierrymoudiki.github.io/blog/2020/04/24/python/r/misc/target-encoder-correlation },
year = { 2020 } }
Previous publications
- Explaining Time-Series Forecasts with Exact Shapley Values (ahead::dynrmf with external regressors applied to scenarios) Mar 8, 2026
- My Presentation at Risk 2026: Lightweight Transfer Learning for Financial Forecasting Mar 1, 2026
- nnetsauce with and without jax for GPU acceleration Feb 23, 2026
- Understanding Boosted Configuration Networks (combined neural networks and boosting): An Intuitive Guide Through Their Hyperparameters Feb 16, 2026
- R version of Python package survivalist, for model-agnostic survival analysis Feb 9, 2026
- Presenting Lightweight Transfer Learning for Financial Forecasting (Risk 2026) Feb 4, 2026
- Option pricing using time series models as market price of risk Feb 1, 2026
- Enhancing Time Series Forecasting (ahead::ridge2f) with Attention-Based Context Vectors (ahead::contextridge2f) Jan 31, 2026
- Overfitting and scaling (on GPU T4) tests on nnetsauce.CustomRegressor Jan 29, 2026
- Beyond Cross-validation: Hyperparameter Optimization via Generalization Gap Modeling Jan 25, 2026
- GPopt for Machine Learning (hyperparameters' tuning) Jan 21, 2026
- rtopy: an R to Python bridge -- novelties Jan 8, 2026
- Python examples for 'Beyond Nelson-Siegel and splines: A model- agnostic Machine Learning framework for discount curve calibration, interpolation and extrapolation' Jan 3, 2026
- Forecasting benchmark: Dynrmf (a new serious competitor in town) vs Theta Method on M-Competitions and Tourism competitition Jan 1, 2026
- Finally figured out a way to port python packages to R using uv and reticulate: example with nnetsauce Dec 17, 2025
- Overfitting Random Fourier Features: Universal Approximation Property Dec 13, 2025
- Counterfactual Scenario Analysis with ahead::ridge2f Dec 11, 2025
- Zero-Shot Probabilistic Time Series Forecasting with TabPFN 2.5 and nnetsauce Dec 10, 2025
- ARIMA Pricing: Semi-Parametric Market price of risk for Risk-Neutral Pricing (code + preprint) Dec 7, 2025
- Analyzing Paper Reviews with LLMs: I Used ChatGPT, DeepSeek, Qwen, Mistral, Gemini, and Claude (and you should too + publish the analysis) Dec 3, 2025
- tisthemachinelearner: New Workflow with uv for R Integration of scikit-learn Dec 1, 2025
- (ICYMI) RPweave: Unified R + Python + LaTeX System using uv Nov 21, 2025
- unifiedml: A Unified Machine Learning Interface for R, is now on CRAN + Discussion about AI replacing humans Nov 16, 2025
- Context-aware Theta forecasting Method: Extending Classical Time Series Forecasting with Machine Learning Nov 13, 2025
- unifiedml in R: A Unified Machine Learning Interface Nov 5, 2025
- Deterministic Shift Adjustment in Arbitrage-Free Pricing (historical to risk-neutral short rates) Oct 28, 2025
- New instantaneous short rates models with their deterministic shift adjustment, for historical and risk-neutral simulation Oct 27, 2025
- RPweave: Unified R + Python + LaTeX System using uv Oct 19, 2025
- GAN-like Synthetic Data Generation Examples (on univariate, multivariate distributions, digits recognition, Fashion-MNIST, stock returns, and Olivetti faces) with DistroSimulator Oct 19, 2025
- R port of llama2.c Oct 9, 2025
- Native uncertainty quantification for time series with NGBoost Oct 8, 2025
- NGBoost (Natural Gradient Boosting) for Regression, Classification, Time Series forecasting and Reserving Oct 6, 2025
- Real-time pricing with a pretrained probabilistic stock return model Oct 1, 2025
- Combining any model with GARCH(1,1) for probabilistic stock forecasting Sep 23, 2025
- Generating Synthetic Data with R-vine Copulas using esgtoolkit in R Sep 21, 2025
- Reimagining Equity Solvency Capital Requirement Approximation (one of my Master's Thesis subjects): From Bilinear Interpolation to Probabilistic Machine Learning Sep 16, 2025
- Transfer Learning using ahead::ridge2f on synthetic stocks returns Pt.2: synthetic data generation Sep 9, 2025
- Transfer Learning using ahead::ridge2f on synthetic stocks returns Sep 8, 2025
- I'm supposed to present 'Conformal Predictive Simulations for Univariate Time Series' at COPA CONFERENCE 2025 in London... Sep 4, 2025
- external regressors in ahead::dynrmf's interface for Machine learning forecasting Sep 1, 2025
- Another interesting decision, now for 'Beyond Nelson-Siegel and splines: A model-agnostic Machine Learning framework for discount curve calibration, interpolation and extrapolation' Aug 20, 2025
- Boosting any randomized based learner for regression, classification and univariate/multivariate time series forcasting Jul 26, 2025
- New nnetsauce version with CustomBackPropRegressor (CustomRegressor with Backpropagation) and ElasticNet2Regressor (Ridge2 with ElasticNet regularization) Jul 15, 2025
- mlsauce (home to a model-agnostic gradient boosting algorithm) can now be installed from PyPI. Jul 10, 2025
- A user-friendly graphical interface to techtonique dot net's API (will eventually contain graphics). Jul 8, 2025
- Calling =TECHTO_MLCLASSIFICATION for Machine Learning supervised CLASSIFICATION in Excel is just a matter of copying and pasting Jul 7, 2025
- Calling =TECHTO_MLREGRESSION for Machine Learning supervised regression in Excel is just a matter of copying and pasting Jul 6, 2025
- Calling =TECHTO_RESERVING and =TECHTO_MLRESERVING for claims triangle reserving in Excel is just a matter of copying and pasting Jul 5, 2025
- Calling =TECHTO_SURVIVAL for Survival Analysis in Excel is just a matter of copying and pasting Jul 4, 2025
- Calling =TECHTO_SIMULATION for Stochastic Simulation in Excel is just a matter of copying and pasting Jul 3, 2025
- Calling =TECHTO_FORECAST for forecasting in Excel is just a matter of copying and pasting Jul 2, 2025
- Random Vector Functional Link (RVFL) artificial neural network with 2 regularization parameters successfully used for forecasting/synthetic simulation in professional settings: Extensions (including Bayesian) Jul 1, 2025
- R version of 'Backpropagating quasi-randomized neural networks' Jun 24, 2025
- Backpropagating quasi-randomized neural networks Jun 23, 2025
- Beyond ARMA-GARCH: leveraging any statistical model for volatility forecasting Jun 21, 2025
- Stacked generalization (Machine Learning model stacking) + conformal prediction for forecasting with ahead::mlf Jun 18, 2025
- An Overfitting dilemma: XGBoost Default Hyperparameters vs GenericBooster + LinearRegression Default Hyperparameters Jun 14, 2025
- Programming language-agnostic reserving using RidgeCV, LightGBM, XGBoost, and ExtraTrees Machine Learning models Jun 13, 2025
- Free R, Python and SQL editors in techtonique dot net Jun 9, 2025
- Beyond Nelson-Siegel and splines: A model-agnostic Machine Learning framework for discount curve calibration, interpolation and extrapolation Jun 7, 2025
- scikit-learn, glmnet, xgboost, lightgbm, pytorch, keras, nnetsauce in probabilistic Machine Learning (for longitudinal data) Reserving (work in progress) Jun 6, 2025
- R version of Probabilistic Machine Learning (for longitudinal data) Reserving (work in progress) Jun 5, 2025
- Probabilistic Machine Learning (for longitudinal data) Reserving (work in progress) Jun 4, 2025
- Python version of Beyond ARMA-GARCH: leveraging model-agnostic Quasi-Randomized networks and conformal prediction for nonparametric probabilistic stock forecasting (ML-ARCH) Jun 3, 2025
- Beyond ARMA-GARCH: leveraging model-agnostic Machine Learning and conformal prediction for nonparametric probabilistic stock forecasting (ML-ARCH) Jun 2, 2025
- Permutations and SHAPley values for feature importance in techtonique dot net's API (with R + Python + the command line) Jun 1, 2025
- Which patient is going to survive longer? Another guide to using techtonique dot net's API (with R + Python + the command line) for survival analysis May 31, 2025
- A Guide to Using techtonique.net's API and rush for simulating and plotting Stochastic Scenarios May 30, 2025
- Simulating Stochastic Scenarios with Diffusion Models: A Guide to Using techtonique.net's API for the purpose May 29, 2025
- Will my apartment in 5th avenue be overpriced or not? Harnessing the power of www.techtonique.net (+ xgboost, lightgbm, catboost) to find out May 28, 2025
- How long must I wait until something happens: A Comprehensive Guide to Survival Analysis via an API May 27, 2025
- Harnessing the Power of techtonique.net: A Comprehensive Guide to Machine Learning Classification via an API May 26, 2025
- Quantile regression with any regressor -- Examples with RandomForestRegressor, RidgeCV, KNeighborsRegressor May 20, 2025
- Survival stacking: survival analysis translated as supervised classification in R and Python May 5, 2025
- 'Bayesian' optimization of hyperparameters in a R machine learning model using the bayesianrvfl package Apr 25, 2025
- A lightweight interface to scikit-learn in R: Bayesian and Conformal prediction Apr 21, 2025
- A lightweight interface to scikit-learn in R Pt.2: probabilistic time series forecasting in conjunction with ahead::dynrmf Apr 20, 2025
- Extending the Theta forecasting method to GLMs, GAMs, GLMBOOST and attention: benchmarking on Tourism, M1, M3 and M4 competition data sets (28000 series) Apr 14, 2025
- Extending the Theta forecasting method to GLMs and attention Apr 8, 2025
- Nonlinear conformalized Generalized Linear Models (GLMs) with R package 'rvfl' (and other models) Mar 31, 2025
- Probabilistic Time Series Forecasting (predictive simulations) in Microsoft Excel using Python, xlwings lite and www.techtonique.net Mar 28, 2025
- Conformalize (improved prediction intervals and simulations) any R Machine Learning model with misc::conformalize Mar 25, 2025
- My poster for the 18th FINANCIAL RISKS INTERNATIONAL FORUM by Institut Louis Bachelier/Fondation du Risque/Europlace Institute of Finance Mar 19, 2025
- Interpretable probabilistic kernel ridge regression using Matérn 3/2 kernels Mar 16, 2025
- (News from) Probabilistic Forecasting of univariate and multivariate Time Series using Quasi-Randomized Neural Networks (Ridge2) and Conformal Prediction Mar 9, 2025
- Word-Online: re-creating Karpathy's char-RNN (with supervised linear online learning of word embeddings) for text completion Mar 8, 2025
- CRAN-like repository for most recent releases of Techtonique's R packages Mar 2, 2025
- Presenting 'Online Probabilistic Estimation of Carbon Beta and Carbon Shapley Values for Financial and Climate Risk' at Institut Louis Bachelier Feb 27, 2025
- Web app with DeepSeek R1 and Hugging Face API for chatting Feb 23, 2025
- tisthemachinelearner: A Lightweight interface to scikit-learn with 2 classes, Classifier and Regressor (in Python and R) Feb 17, 2025
- R version of survivalist: Probabilistic model-agnostic survival analysis using scikit-learn, xgboost, lightgbm (and conformal prediction) Feb 12, 2025
- Model-agnostic global Survival Prediction of Patients with Myeloid Leukemia in QRT/Gustave Roussy Challenge (challengedata.ens.fr): Python's survivalist Quickstart Feb 10, 2025
- A simple test of the martingale hypothesis in esgtoolkit Feb 3, 2025
- Command Line Interface (CLI) for techtonique.net's API Jan 31, 2025
- Gradient-Boosting and Boostrap aggregating anything (alert: high performance): Part5, easier install and Rust backend Jan 27, 2025
- Just got a paper on conformal prediction REJECTED by International Journal of Forecasting despite evidence on 30,000 time series (and more). What's going on? Part2: 1311 time series from the Tourism competition Jan 20, 2025
- Techtonique is out! (with a tutorial in various programming languages and formats) Jan 14, 2025
- Univariate and Multivariate Probabilistic Forecasting with nnetsauce and TabPFN Jan 14, 2025
- Just got a paper on conformal prediction REJECTED by International Journal of Forecasting despite evidence on 30,000 time series (and more). What's going on? Jan 5, 2025
- Python and Interactive dashboard version of Stock price forecasting with Deep Learning: throwing power at the problem (and why it won't make you rich) Dec 31, 2024
- Stock price forecasting with Deep Learning: throwing power at the problem (and why it won't make you rich) Dec 29, 2024
- No-code Machine Learning Cross-validation and Interpretability in techtonique.net Dec 23, 2024
- survivalist: Probabilistic model-agnostic survival analysis using scikit-learn, glmnet, xgboost, lightgbm, pytorch, keras, nnetsauce and mlsauce Dec 15, 2024
- Model-agnostic 'Bayesian' optimization (for hyperparameter tuning) using conformalized surrogates in GPopt Dec 9, 2024
- You can beat Forecasting LLMs (Large Language Models a.k.a foundation models) with nnetsauce.MTS Pt.2: Generic Gradient Boosting Dec 1, 2024
- You can beat Forecasting LLMs (Large Language Models a.k.a foundation models) with nnetsauce.MTS Nov 24, 2024
- Unified interface and conformal prediction (calibrated prediction intervals) for R package forecast (and 'affiliates') Nov 23, 2024
- GLMNet in Python: Generalized Linear Models Nov 18, 2024
- Gradient-Boosting anything (alert: high performance): Part4, Time series forecasting Nov 10, 2024
- Predictive scenarios simulation in R, Python and Excel using Techtonique API Nov 3, 2024
- Chat with your tabular data in www.techtonique.net Oct 30, 2024
- Gradient-Boosting anything (alert: high performance): Part3, Histogram-based boosting Oct 28, 2024
- R editor and SQL console (in addition to Python editors) in www.techtonique.net Oct 21, 2024
- R and Python consoles + JupyterLite in www.techtonique.net Oct 15, 2024
- Gradient-Boosting anything (alert: high performance): Part2, R version Oct 14, 2024
- Gradient-Boosting anything (alert: high performance) Oct 6, 2024
- Benchmarking 30 statistical/Machine Learning models on the VN1 Forecasting -- Accuracy challenge Oct 4, 2024
- Automated random variable distribution inference using Kullback-Leibler divergence and simulating best-fitting distribution Oct 2, 2024
- Forecasting in Excel using Techtonique's Machine Learning APIs under the hood Sep 30, 2024
- Techtonique web app for data-driven decisions using Mathematics, Statistics, Machine Learning, and Data Visualization Sep 25, 2024
- Parallel for loops (Map or Reduce) + New versions of nnetsauce and ahead Sep 16, 2024
- Adaptive (online/streaming) learning with uncertainty quantification using Polyak averaging in learningmachine Sep 10, 2024
- New versions of nnetsauce and ahead Sep 9, 2024
- Prediction sets and prediction intervals for conformalized Auto XGBoost, Auto LightGBM, Auto CatBoost, Auto GradientBoosting Sep 2, 2024
- Quick/automated R package development workflow (assuming you're using macOS or Linux) Part2 Aug 30, 2024
- R package development workflow (assuming you're using macOS or Linux) Aug 27, 2024
- A new method for deriving a nonparametric confidence interval for the mean Aug 26, 2024
- Conformalized adaptive (online/streaming) learning using learningmachine in Python and R Aug 19, 2024
- Bayesian (nonlinear) adaptive learning Aug 12, 2024
- Auto XGBoost, Auto LightGBM, Auto CatBoost, Auto GradientBoosting Aug 5, 2024
- Copulas for uncertainty quantification in time series forecasting Jul 28, 2024
- Forecasting uncertainty: sequential split conformal prediction + Block bootstrap (web app) Jul 22, 2024
- learningmachine for Python (new version) Jul 15, 2024
- learningmachine v2.0.0: Machine Learning with explanations and uncertainty quantification Jul 8, 2024
- My presentation at ISF 2024 conference (slides with nnetsauce probabilistic forecasting news) Jul 3, 2024
- 10 uncertainty quantification methods in nnetsauce forecasting Jul 1, 2024
- Forecasting with XGBoost embedded in Quasi-Randomized Neural Networks Jun 24, 2024
- Forecasting Monthly Airline Passenger Numbers with Quasi-Randomized Neural Networks Jun 17, 2024
- Automated hyperparameter tuning using any conformalized surrogate Jun 9, 2024
- Recognizing handwritten digits with Ridge2Classifier Jun 3, 2024
- Forecasting the Economy May 27, 2024
- A detailed introduction to Deep Quasi-Randomized 'neural' networks May 19, 2024
- Probability of receiving a loan; using learningmachine May 12, 2024
- mlsauce's `v0.18.2`: various examples and benchmarks with dimension reduction May 6, 2024
- mlsauce's `v0.17.0`: boosting with Elastic Net, polynomials and heterogeneity in explanatory variables Apr 29, 2024
- mlsauce's `v0.13.0`: taking into account inputs heterogeneity through clustering Apr 21, 2024
- mlsauce's `v0.12.0`: prediction intervals for LSBoostRegressor Apr 15, 2024
- Conformalized predictive simulations for univariate time series on more than 250 data sets Apr 7, 2024
- learningmachine v1.1.2: for Python Apr 1, 2024
- learningmachine v1.0.0: prediction intervals around the probability of the event 'a tumor being malignant' Mar 25, 2024
- Bayesian inference and conformal prediction (prediction intervals) in nnetsauce v0.18.1 Mar 18, 2024
- Multiple examples of Machine Learning forecasting with ahead Mar 11, 2024
- rtopy (v0.1.1): calling R functions in Python Mar 4, 2024
- ahead forecasting (v0.10.0): fast time series model calibration and Python plots Feb 26, 2024
- A plethora of datasets at your fingertips Part3: how many times do couples cheat on each other? Feb 19, 2024
- nnetsauce's introduction as of 2024-02-11 (new version 0.17.0) Feb 11, 2024
- Tuning Machine Learning models with GPopt's new version Part 2 Feb 5, 2024
- Tuning Machine Learning models with GPopt's new version Jan 29, 2024
- Subsampling continuous and discrete response variables Jan 22, 2024
- DeepMTS, a Deep Learning Model for Multivariate Time Series Jan 15, 2024
- A classifier that's very accurate (and deep) Pt.2: there are > 90 classifiers in nnetsauce Jan 8, 2024
- learningmachine: prediction intervals for conformalized Kernel ridge regression and Random Forest Jan 1, 2024
- A plethora of datasets at your fingertips Part2: how many times do couples cheat on each other? Descriptive analytics, interpretability and prediction intervals using conformal prediction Dec 25, 2023
- Diffusion models in Python with esgtoolkit (Part2) Dec 18, 2023
- Diffusion models in Python with esgtoolkit Dec 11, 2023
- Julia packaging at the command line Dec 4, 2023
- Quasi-randomized nnetworks in Julia, Python and R Nov 27, 2023
- A plethora of datasets at your fingertips Nov 20, 2023
- A classifier that's very accurate (and deep) Nov 12, 2023
- mlsauce version 0.8.10: Statistical/Machine Learning with Python and R Nov 5, 2023
- AutoML in nnetsauce (randomized and quasi-randomized nnetworks) Pt.2: multivariate time series forecasting Oct 29, 2023
- AutoML in nnetsauce (randomized and quasi-randomized nnetworks) Oct 22, 2023
- Version v0.14.0 of nnetsauce for R and Python Oct 16, 2023
- A diffusion model: G2++ Oct 9, 2023
- Diffusion models in ESGtoolkit + announcements Oct 2, 2023
- An infinity of time series forecasting models in nnetsauce (Part 2 with uncertainty quantification) Sep 25, 2023
- (News from) forecasting in Python with ahead (progress bars and plots) Sep 18, 2023
- Forecasting in Python with ahead Sep 11, 2023
- Risk-neutralize simulations Sep 4, 2023
- Comparing cross-validation results using crossval_ml and boxplots Aug 27, 2023
- Reminder Apr 30, 2023
- Did you ask ChatGPT about who you are? Apr 16, 2023
- A new version of nnetsauce (randomized and quasi-randomized 'neural' networks) Apr 2, 2023
- Simple interfaces to the forecasting API Nov 23, 2022
- A web application for forecasting in Python, R, Ruby, C#, JavaScript, PHP, Go, Rust, Java, MATLAB, etc. Nov 2, 2022
- Prediction intervals (not only) for Boosted Configuration Networks in Python Oct 5, 2022
- Boosted Configuration (neural) Networks Pt. 2 Sep 3, 2022
- Boosted Configuration (_neural_) Networks for classification Jul 21, 2022
- A Machine Learning workflow using Techtonique Jun 6, 2022
- Super Mario Bros © in the browser using PyScript May 8, 2022
- News from ESGtoolkit, ycinterextra, and nnetsauce Apr 4, 2022
- Explaining a Keras _neural_ network predictions with the-teller Mar 11, 2022
- New version of nnetsauce -- various quasi-randomized networks Feb 12, 2022
- A dashboard illustrating bivariate time series forecasting with `ahead` Jan 14, 2022
- Hundreds of Statistical/Machine Learning models for univariate time series, using ahead, ranger, xgboost, and caret Dec 20, 2021
- Forecasting with `ahead` (Python version) Dec 13, 2021
- Tuning and interpreting LSBoost Nov 15, 2021
- Time series cross-validation using `crossvalidation` (Part 2) Nov 7, 2021
- Fast and scalable forecasting with ahead::ridge2f Oct 31, 2021
- Automatic Forecasting with `ahead::dynrmf` and Ridge regression Oct 22, 2021
- Forecasting with `ahead` Oct 15, 2021
- Classification using linear regression Sep 26, 2021
- `crossvalidation` and random search for calibrating support vector machines Aug 6, 2021
- parallel grid search cross-validation using `crossvalidation` Jul 31, 2021
- `crossvalidation` on R-universe, plus a classification example Jul 23, 2021
- Documentation and source code for GPopt, a package for Bayesian optimization Jul 2, 2021
- Hyperparameters tuning with GPopt Jun 11, 2021
- A forecasting tool (API) with examples in curl, R, Python May 28, 2021
- Bayesian Optimization with GPopt Part 2 (save and resume) Apr 30, 2021
- Bayesian Optimization with GPopt Apr 16, 2021
- Compatibility of nnetsauce and mlsauce with scikit-learn Mar 26, 2021
- Explaining xgboost predictions with the teller Mar 12, 2021
- An infinity of time series models in nnetsauce Mar 6, 2021
- New activation functions in mlsauce's LSBoost Feb 12, 2021
- 2020 recap, Gradient Boosting, Generalized Linear Models, AdaOpt with nnetsauce and mlsauce Dec 29, 2020
- A deeper learning architecture in nnetsauce Dec 18, 2020
- Classify penguins with nnetsauce's MultitaskClassifier Dec 11, 2020
- Bayesian forecasting for uni/multivariate time series Dec 4, 2020
- Generalized nonlinear models in nnetsauce Nov 28, 2020
- Boosting nonlinear penalized least squares Nov 21, 2020
- Statistical/Machine Learning explainability using Kernel Ridge Regression surrogates Nov 6, 2020
- NEWS Oct 30, 2020
- A glimpse into my PhD journey Oct 23, 2020
- Submitting R package to CRAN Oct 16, 2020
- Simulation of dependent variables in ESGtoolkit Oct 9, 2020
- Forecasting lung disease progression Oct 2, 2020
- New nnetsauce Sep 25, 2020
- Technical documentation Sep 18, 2020
- A new version of nnetsauce, and a new Techtonique website Sep 11, 2020
- Back next week, and a few announcements Sep 4, 2020
- Explainable 'AI' using Gradient Boosted randomized networks Pt2 (the Lasso) Jul 31, 2020
- LSBoost: Explainable 'AI' using Gradient Boosted randomized networks (with examples in R and Python) Jul 24, 2020
- nnetsauce version 0.5.0, randomized neural networks on GPU Jul 17, 2020
- Maximizing your tip as a waiter (Part 2) Jul 10, 2020
- New version of mlsauce, with Gradient Boosted randomized networks and stump decision trees Jul 3, 2020
- Announcements Jun 26, 2020
- Parallel AdaOpt classification Jun 19, 2020
- Comments section and other news Jun 12, 2020
- Maximizing your tip as a waiter Jun 5, 2020
- AdaOpt classification on MNIST handwritten digits (without preprocessing) May 29, 2020
- AdaOpt (a probabilistic classifier based on a mix of multivariable optimization and nearest neighbors) for R May 22, 2020
- AdaOpt May 15, 2020
- Custom errors for cross-validation using crossval::crossval_ml May 8, 2020
- Documentation+Pypi for the `teller`, a model-agnostic tool for Machine Learning explainability May 1, 2020
- Encoding your categorical variables based on the response variable and correlations Apr 24, 2020
- Linear model, xgboost and randomForest cross-validation using crossval::crossval_ml Apr 17, 2020
- Grid search cross-validation using crossval Apr 10, 2020
- Documentation for the querier, a query language for Data Frames Apr 3, 2020
- Time series cross-validation using crossval Mar 27, 2020
- On model specification, identification, degrees of freedom and regularization Mar 20, 2020
- Import data into the querier (now on Pypi), a query language for Data Frames Mar 13, 2020
- R notebooks for nnetsauce Mar 6, 2020
- Version 0.4.0 of nnetsauce, with fruits and breast cancer classification Feb 28, 2020
- Create a specific feed in your Jekyll blog Feb 21, 2020
- Git/Github for contributing to package development Feb 14, 2020
- Feedback forms for contributing Feb 7, 2020
- nnetsauce for R Jan 31, 2020
- A new version of nnetsauce (v0.3.1) Jan 24, 2020
- ESGtoolkit, a tool for Monte Carlo simulation (v0.2.0) Jan 17, 2020
- Search bar, new year 2020 Jan 10, 2020
- 2019 Recap, the nnetsauce, the teller and the querier Dec 20, 2019
- Understanding model interactions with the `teller` Dec 13, 2019
- Using the `teller` on a classifier Dec 6, 2019
- Benchmarking the querier's verbs Nov 29, 2019
- Composing the querier's verbs for data wrangling Nov 22, 2019
- Comparing and explaining model predictions with the teller Nov 15, 2019
- Tests for the significance of marginal effects in the teller Nov 8, 2019
- Introducing the teller Nov 1, 2019
- Introducing the querier Oct 25, 2019
- Prediction intervals for nnetsauce models Oct 18, 2019
- Using R in Python for statistical learning/data science Oct 11, 2019
- Model calibration with `crossval` Oct 4, 2019
- Bagging in the nnetsauce Sep 25, 2019
- Adaboost learning with nnetsauce Sep 18, 2019
- Change in blog's presentation Sep 4, 2019
- nnetsauce on Pypi Jun 5, 2019
- More nnetsauce (examples of use) May 9, 2019
- nnetsauce Mar 13, 2019
- crossval Mar 13, 2019
- test Mar 10, 2019


Comments powered by Talkyard.