Today, give a try to Techtonique web app, a tool designed to help you make informed, data-driven decisions using Mathematics, Statistics, Machine Learning, and Data Visualization. Here is a tutorial with audio, video, code, and slides: https://moudiki2.gumroad.com/l/nrhgb. 100 API requests are now (and forever) offered to every user every month, no matter the pricing tier.
Introduction
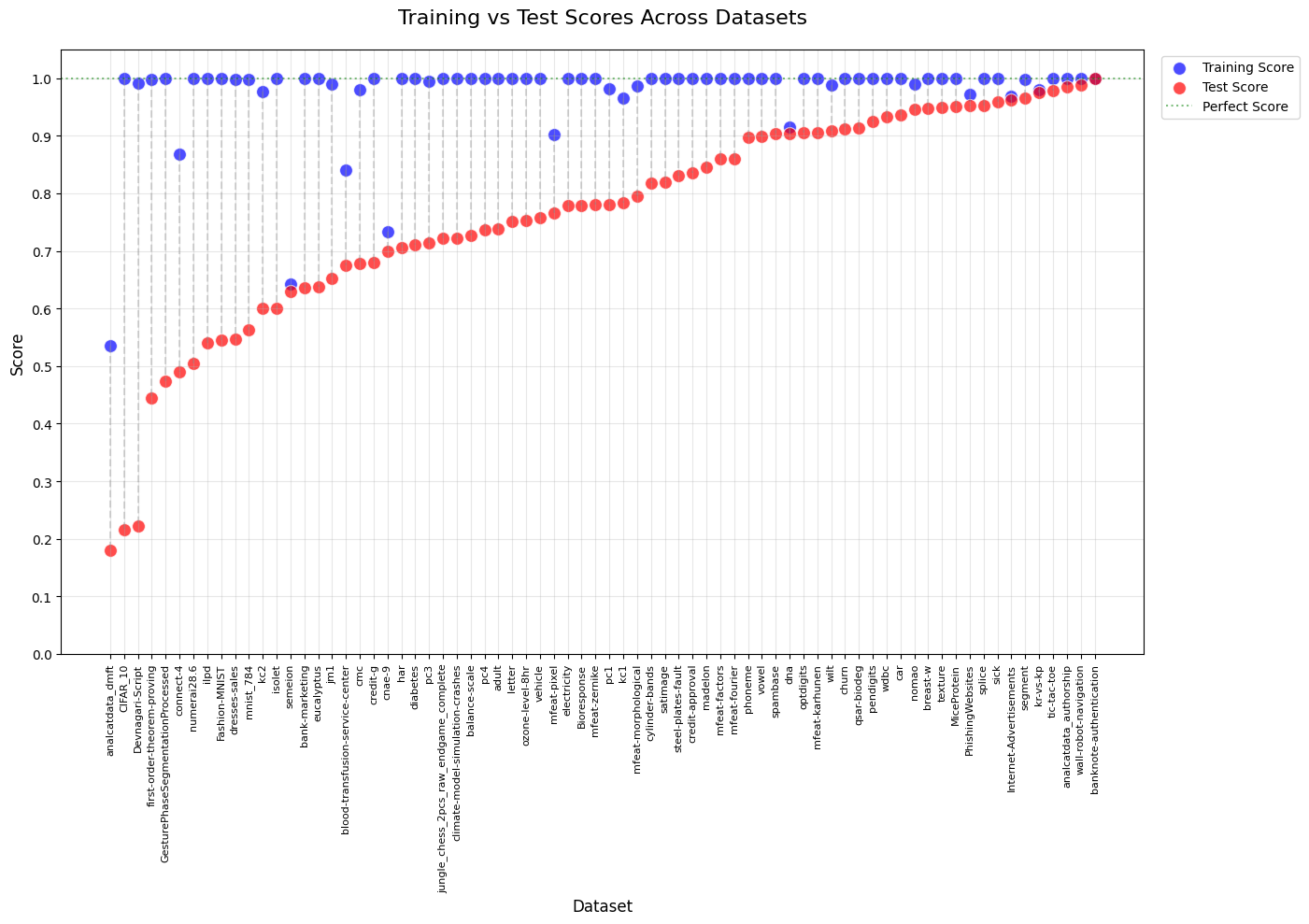
In this post, we’ll explore an intriguing comparison between XGBoost’s balanced accuracy with its default hyperparameters on 72 data sets and a GenericBooster with Linear Regression as a base learner, also with its default hyperparameters. Whereas the first one clearly overfits when not tuned (to an extent that I wanted to verify), but gives an overall higher test set balanced accuracy, the second one gives highly correlated training and test set errors, with an overall lower test set balanced accuracy.
What We’ll Cover
- Setting up the environment with necessary packages
- Loading and preparing the data
- Comparing model performances
- Analyzing overfitting patterns
- Drawing conclusions about which approach might be better for specific scenarios
Let’s dive into the implementation and analysis.
!pip install genbooster
!pip install nnetsauce
Import the necessary packages.
import xgboost as xgb
import joblib
import requests
import io
from genbooster.genboosterclassifier import BoosterClassifier
from genbooster.randombagclassifier import RandomBagClassifier
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import BaggingRegressor
from sklearn.linear_model import Ridge, LinearRegression
from sklearn.gaussian_process import GaussianProcessRegressor
from tqdm import tqdm
from scipy import stats
from sklearn.model_selection import train_test_split
from sklearn.metrics import balanced_accuracy_score
from sklearn.model_selection import cross_val_score
from sklearn.metrics import make_scorer
from time import time
Obtain 72 data sets, a reduced form of the openml-cc18, available at https://github.com/thierrymoudiki/openml-cc18-reduced.
# Fetch the file content from the URL - **Use the raw file URL!**
url = 'https://github.com/thierrymoudiki/openml-cc18-reduced/raw/main/openml-cc18-Xys-2024-05-20.pkl'
# Use a session to handle potential connection interruptions
session = requests.Session()
response = session.get(url, stream=True)
response.raise_for_status() # Raise an exception for bad responses
# Load the data from the downloaded content in chunks
with io.BytesIO() as buffer:
for chunk in response.iter_content(chunk_size=1024*1024): # Download in 1MB chunks
buffer.write(chunk)
buffer.seek(0) # Reset buffer position to the beginning
clf_datasets = joblib.load(buffer)
1 - xgboost results
results = {}
balanced_accuracy_scorer = make_scorer(balanced_accuracy_score)
for i, dataset in tqdm(enumerate(clf_datasets.items())):
dataset_name = dataset[0]
print("\n ----------", (i + 1), "/", len(clf_datasets.items()), ": ", dataset_name)
try:
X, y = dataset[1]['dataset'][0], dataset[1]['dataset'][1]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42, stratify=y)
# Split dataset into training and testing sets
model = xgb.XGBClassifier()
model.fit(X_train, y_train)
y_pred_train = model.predict(X_train)
y_pred = model.predict(X_test)
# Calculate balanced accuracy
balanced_acc_train = balanced_accuracy_score(y_train, y_pred_train)
balanced_acc_test = balanced_accuracy_score(y_test, y_pred)
print("Training Balanced Accuracy:", balanced_acc_train)
print("Testing Balanced Accuracy:", balanced_acc_test)
results[dataset_name] = {"training_score": balanced_acc_train,
"testing_score": balanced_acc_test}
except:
print("Error")
0it [00:00, ?it/s]
---------- 1 / 72 : kr-vs-kp
1it [00:00, 4.01it/s]
Training Balanced Accuracy: 0.9793965874420882
Testing Balanced Accuracy: 0.975187969924812
---------- 2 / 72 : letter
2it [00:03, 2.19s/it]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.7513736263736264
---------- 3 / 72 : balance-scale
3it [00:04, 1.52s/it]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.7260334744908249
---------- 4 / 72 : mfeat-factors
4it [00:09, 2.97s/it]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.86
---------- 5 / 72 : mfeat-fourier
5it [00:16, 4.19s/it]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.86
---------- 6 / 72 : breast-w
6it [00:17, 3.20s/it]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.9470108695652174
---------- 7 / 72 : mfeat-karhunen
7it [00:22, 3.84s/it]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.905
---------- 8 / 72 : mfeat-morphological
8it [00:22, 2.76s/it]
Training Balanced Accuracy: 0.9862499999999998
Testing Balanced Accuracy: 0.7950000000000002
---------- 9 / 72 : mfeat-zernike
10it [00:23, 1.46s/it]
Training Balanced Accuracy: 0.99875
Testing Balanced Accuracy: 0.7799999999999999
---------- 10 / 72 : cmc
Training Balanced Accuracy: 0.9798009974256786
Testing Balanced Accuracy: 0.6773695839418791
---------- 11 / 72 : optdigits
13it [00:23, 1.66it/s]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.905
---------- 12 / 72 : credit-approval
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.8353204172876304
---------- 13 / 72 : credit-g
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.6797619047619048
---------- 14 / 72 : pendigits
16it [00:24, 3.01it/s]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.9240601503759398
---------- 15 / 72 : diabetes
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.7112962962962963
---------- 16 / 72 : spambase
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.9041740767862747
---------- 17 / 72 : splice
18it [00:24, 4.30it/s]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.9529914529914528
---------- 18 / 72 : tic-tac-toe
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.9776119402985075
---------- 19 / 72 : vehicle
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.7570075757575757
---------- 20 / 72 : electricity
21it [00:24, 5.32it/s]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.778005115089514
---------- 21 / 72 : satimage
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.8195766960114664
---------- 22 / 72 : eucalyptus
22it [00:25, 5.05it/s]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.6379919821780285
---------- 23 / 72 : sick
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.9583333333333333
---------- 24 / 72 : vowel
24it [00:25, 4.56it/s]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.898989898989899
---------- 25 / 72 : isolet
25it [00:26, 2.19it/s]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.6002747252747253
---------- 26 / 72 : analcatdata_authorship
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.9850446428571429
---------- 27 / 72 : analcatdata_dmft
27it [00:27, 2.97it/s]
Training Balanced Accuracy: 0.5354622066211941
Testing Balanced Accuracy: 0.1808442851453604
---------- 28 / 72 : mnist_784
30it [00:27, 3.67it/s]
Training Balanced Accuracy: 0.9974358974358974
Testing Balanced Accuracy: 0.5634728124659475
---------- 29 / 72 : pc4
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.7357954545454546
---------- 30 / 72 : pc3
Training Balanced Accuracy: 0.9939024390243902
Testing Balanced Accuracy: 0.7138888888888889
---------- 31 / 72 : jm1
33it [00:28, 5.62it/s]
Training Balanced Accuracy: 0.9894845464612907
Testing Balanced Accuracy: 0.6519350215002389
---------- 32 / 72 : kc2
Training Balanced Accuracy: 0.9764705882352941
Testing Balanced Accuracy: 0.6002190580503833
---------- 33 / 72 : kc1
Training Balanced Accuracy: 0.9660003848559195
Testing Balanced Accuracy: 0.7827829738499714
---------- 34 / 72 : pc1
35it [00:28, 7.30it/s]
Training Balanced Accuracy: 0.9818181818181818
Testing Balanced Accuracy: 0.7803379416282642
---------- 35 / 72 : adult
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.7384868421052632
---------- 36 / 72 : Bioresponse
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.7790603891521323
---------- 37 / 72 : wdbc
39it [00:28, 9.19it/s]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.933531746031746
---------- 38 / 72 : phoneme
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.8967423969227071
---------- 39 / 72 : qsar-biodeg
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.9139827179890023
---------- 40 / 72 : wall-robot-navigation
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.9885802469135803
---------- 41 / 72 : semeion
43it [00:29, 8.21it/s]
Training Balanced Accuracy: 0.6431816659664762
Testing Balanced Accuracy: 0.63
---------- 42 / 72 : ilpd
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.5405740609496811
---------- 43 / 72 : madelon
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.845
---------- 44 / 72 : nomao
45it [00:29, 9.50it/s]
Training Balanced Accuracy: 0.9890350877192983
Testing Balanced Accuracy: 0.9456508403876824
---------- 45 / 72 : ozone-level-8hr
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.7531879884821061
---------- 46 / 72 : cnae-9
Training Balanced Accuracy: 0.7333304959709455
Testing Balanced Accuracy: 0.699604743083004
---------- 47 / 72 : first-order-theorem-proving
50it [00:30, 7.28it/s]
Training Balanced Accuracy: 0.9974161661661661
Testing Balanced Accuracy: 0.44510582010582017
---------- 48 / 72 : banknote-authentication
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 1.0
---------- 49 / 72 : blood-transfusion-service-center
Training Balanced Accuracy: 0.8398196194712132
Testing Balanced Accuracy: 0.6754385964912281
---------- 50 / 72 : PhishingWebsites
Training Balanced Accuracy: 0.9715260585285342
Testing Balanced Accuracy: 0.9516145358841988
---------- 51 / 72 : cylinder-bands
52it [00:30, 8.60it/s]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.8183730715287517
---------- 52 / 72 : bank-marketing
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.6352247605011054
---------- 53 / 72 : GesturePhaseSegmentationProcessed
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.473961038961039
---------- 54 / 72 : har
54it [00:31, 4.44it/s]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.7054457054457054
---------- 55 / 72 : dresses-sales
Training Balanced Accuracy: 0.9970238095238095
Testing Balanced Accuracy: 0.5472085385878489
---------- 56 / 72 : texture
57it [00:31, 5.19it/s]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.9494949494949495
---------- 57 / 72 : connect-4
Training Balanced Accuracy: 0.8687185238384995
Testing Balanced Accuracy: 0.490677451203767
---------- 58 / 72 : MiceProtein
58it [00:32, 4.66it/s]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.9510996240601504
---------- 59 / 72 : steel-plates-fault
59it [00:32, 3.88it/s]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.8302055604850634
---------- 60 / 72 : climate-model-simulation-crashes
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.7222222222222222
---------- 61 / 72 : wilt
Training Balanced Accuracy: 0.9880952380952381
Testing Balanced Accuracy: 0.9090909090909092
---------- 62 / 72 : car
62it [00:32, 5.28it/s]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.9357142857142857
---------- 63 / 72 : segment
63it [00:34, 1.90it/s]
Training Balanced Accuracy: 0.9974826446647592
Testing Balanced Accuracy: 0.9649894440534835
---------- 64 / 72 : mfeat-pixel
64it [00:35, 2.15it/s]
Training Balanced Accuracy: 0.9012499999999999
Testing Balanced Accuracy: 0.7649999999999999
---------- 65 / 72 : Fashion-MNIST
66it [00:36, 2.41it/s]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.545
---------- 66 / 72 : jungle_chess_2pcs_raw_endgame_complete
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.7222113037136032
---------- 67 / 72 : numerai28.6
67it [00:36, 2.87it/s]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.5044004400440043
---------- 68 / 72 : Devnagari-Script
68it [00:38, 1.32it/s]
Training Balanced Accuracy: 0.9909686700767264
Testing Balanced Accuracy: 0.2217391304347826
---------- 69 / 72 : CIFAR_10
71it [00:39, 2.08it/s]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.215
---------- 70 / 72 : Internet-Advertisements
Training Balanced Accuracy: 0.967741724282422
Testing Balanced Accuracy: 0.9613787375415282
---------- 71 / 72 : dna
Training Balanced Accuracy: 0.9147506693440429
Testing Balanced Accuracy: 0.9043803418803419
---------- 72 / 72 : churn
72it [00:39, 1.84it/s]
Training Balanced Accuracy: 1.0
Testing Balanced Accuracy: 0.9111295681063123
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
import numpy as np
# Your data
data = results
# Convert to DataFrame - correct approach
df = pd.DataFrame.from_dict({(i,j): data[i][j]
for i in data.keys()
for j in data[i].keys()},
orient='index')
# The index is a MultiIndex with (dataset, score_type)
# Let's properly unpack it
df = df.reset_index()
df.columns = ['index', 'score'] # Temporary column names
# Split the multi-index into separate columns
df[['dataset', 'score_type']] = pd.DataFrame(df['index'].tolist(), index=df.index)
df = df.drop(columns=['index'])
# Now pivot the data
plot_df = df.pivot(index='dataset', columns='score_type', values='score')
# Sort by test score for better visualization
plot_df = plot_df.sort_values('testing_score')
# Create figure
plt.figure(figsize=(14, 10), dpi=100)
# Plot training and test scores
sns.scatterplot(data=plot_df, x=plot_df.index, y='training_score',
color='blue', label='Training Score', s=100, alpha=0.7)
sns.scatterplot(data=plot_df, x=plot_df.index, y='testing_score',
color='red', label='Test Score', s=100, alpha=0.7)
# Add connecting lines
for i, dataset in enumerate(plot_df.index):
plt.plot([i, i], [plot_df.loc[dataset, 'training_score'],
plot_df.loc[dataset, 'testing_score']],
color='gray', alpha=0.3, linestyle='--')
# Add perfect score line
plt.axhline(1.0, color='green', linestyle=':', alpha=0.5, label='Perfect Score')
# Customize plot
plt.title('Training vs Test Scores Across Datasets', fontsize=16, pad=20)
plt.xlabel('Dataset', fontsize=12)
plt.ylabel('Score', fontsize=12)
plt.xticks(rotation=90, fontsize=8)
plt.yticks(np.arange(0, 1.1, 0.1))
plt.ylim(0, 1.05)
plt.legend(loc='upper right', bbox_to_anchor=(1.15, 1))
plt.grid(True, alpha=0.3)
# Adjust layout
plt.tight_layout()
plt.show()
import numpy as np
import pandas as pd
from scipy.stats import ttest_rel, wilcoxon, shapiro
import matplotlib.pyplot as plt
import seaborn as sns
def detect_overfitting(train_scores, test_scores, alpha=0.05, plot=True):
"""
Performs statistical tests to detect overfitting between training and test scores,
including BOTH paired t-test and Wilcoxon signed-rank test for robustness.
Parameters:
-----------
train_scores : array-like
Array of training scores (e.g., accuracies) for multiple datasets
test_scores : array-like
Array of test scores for the same datasets
alpha : float, default=0.05
Significance level for statistical tests
plot : bool, default=True
Whether to generate diagnostic plots
Returns:
--------
dict
Dictionary containing test results and effect sizes
"""
# Convert to numpy arrays
train_scores = np.asarray(train_scores)
test_scores = np.asarray(test_scores)
differences = train_scores - test_scores
# Initialize results dictionary
results = {
'n_datasets': len(train_scores),
'mean_train_score': np.mean(train_scores),
'mean_test_score': np.mean(test_scores),
'mean_difference': np.mean(differences),
'median_difference': np.median(differences),
'std_difference': np.std(differences, ddof=1),
}
# Normality test (for interpretation guidance)
_, shapiro_p = shapiro(differences)
results['normality_shapiro_p'] = shapiro_p
results['normal_distribution'] = shapiro_p >= alpha
# ========================
# 1. Paired t-test (parametric)
# ========================
t_stat, t_p = ttest_rel(train_scores, test_scores, alternative='greater')
results['paired_ttest'] = {
'statistic': t_stat,
'p_value': t_p,
'significant': t_p < alpha
}
# ========================
# 2. Wilcoxon signed-rank test (non-parametric)
# ========================
w_stat, w_p = wilcoxon(train_scores, test_scores, alternative='greater')
results['wilcoxon_test'] = {
'statistic': w_stat,
'p_value': w_p,
'significant': w_p < alpha
}
# ========================
# Effect size measures
# ========================
# Cohen's d (standardized mean difference)
results['cohens_d'] = results['mean_difference'] / results['std_difference'] if results['std_difference'] > 0 else 0
# Rank-biserial correlation (for Wilcoxon)
n = len(differences)
results['rank_biserial'] = 1 - (2 * w_stat) / (n * (n + 1)) if n > 0 else 0
# Effect size classification
for eff_size in ['cohens_d', 'rank_biserial']:
val = abs(results[eff_size])
if val > 0.8:
results[f'{eff_size}_interpret'] = 'large'
elif val > 0.5:
results[f'{eff_size}_interpret'] = 'medium'
else:
results[f'{eff_size}_interpret'] = 'small'
# ========================
# Generate plots
# ========================
if plot:
plt.figure(figsize=(15, 5))
# Plot 1: Training vs Test scores
plt.subplot(1, 3, 1)
sns.scatterplot(x=train_scores, y=test_scores)
plt.plot([0, 1], [0, 1], 'k--', alpha=0.5)
plt.xlabel('Training Scores')
plt.ylabel('Test Scores')
plt.title('Training vs Test Scores')
plt.grid(True, alpha=0.3)
# Plot 2: Distribution of differences
plt.subplot(1, 3, 2)
sns.histplot(differences, kde=True)
plt.axvline(results['mean_difference'], color='r', linestyle='--', label='Mean')
plt.axvline(results['median_difference'], color='g', linestyle=':', label='Median')
plt.xlabel('Train Score - Test Score')
plt.title('Distribution of Differences')
plt.legend()
plt.grid(True, alpha=0.3)
# Plot 3: Boxplot of scores
plt.subplot(1, 3, 3)
pd.DataFrame({
'Training': train_scores,
'Test': test_scores
}).boxplot()
plt.title('Score Distributions')
plt.grid(True, alpha=0.3)
plt.tight_layout()
plt.show()
return results
# Run the analysis
training_scores = [result['training_score'] for result in results.values()]
testing_scores = [result['testing_score'] for result in results.values()]
results = detect_overfitting(training_scores, testing_scores)
# Print results in a readable format
print("=== Overfitting Detection Report ===")
print(f"Datasets: {results['n_datasets']}")
print(f"Mean training score: {results['mean_train_score']:.3f}")
print(f"Mean test score: {results['mean_test_score']:.3f}")
print(f"Mean difference: {results['mean_difference']:.3f} (SD={results['std_difference']:.3f})")
print(f"Spearman rho: {stats.spearmanr(training_scores, testing_scores).correlation:.3f}")
print("\n=== Normality Test ===")
print(f"Shapiro-Wilk p-value: {results['normality_shapiro_p']:.4f}")
print("Interpretation:", "Normal distribution" if results['normal_distribution'] else "Non-normal distribution")
print("\n=== Statistical Tests ===")
print("1. Paired t-test:")
print(f" t = {results['paired_ttest']['statistic']:.3f}, p = {results['paired_ttest']['p_value']:.5f}")
print(f" Significant: {'✅' if results['paired_ttest']['significant'] else '❌'}")
print("\n2. Wilcoxon signed-rank test:")
print(f" W = {results['wilcoxon_test']['statistic']:.3f}, p = {results['wilcoxon_test']['p_value']:.5f}")
print(f" Significant: {'✅' if results['wilcoxon_test']['significant'] else '❌'}")
print("\n=== Effect Sizes ===")
print(f"Cohen's d: {results['cohens_d']:.3f} ({results['cohens_d_interpret']} effect)")
print(f"Rank-biserial correlation: {results['rank_biserial']:.3f} ({results['rank_biserial_interpret']} effect)")
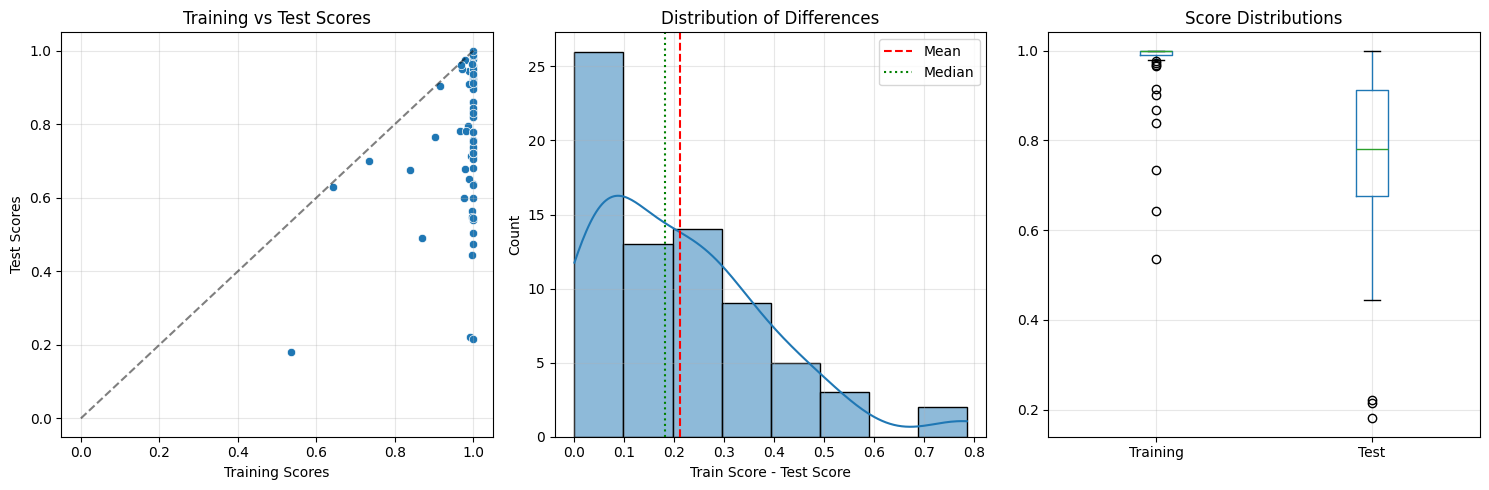
=== Overfitting Detection Report ===
Datasets: 72
Mean training score: 0.975
Mean test score: 0.764
Mean difference: 0.210 (SD=0.175)
Spearman rho: 0.208
=== Normality Test ===
Shapiro-Wilk p-value: 0.0001
Interpretation: Non-normal distribution
=== Statistical Tests ===
1. Paired t-test:
t = 10.184, p = 0.00000
Significant: ✅
2. Wilcoxon signed-rank test:
W = 2556.000, p = 0.00000
Significant: ✅
=== Effect Sizes ===
Cohen's d: 1.200 (large effect)
Rank-biserial correlation: 0.027 (small effect)
2 - Genbooster
from os import name
from sklearn.linear_model import LinearRegression
from sklearn.tree import DecisionTreeRegressor, ExtraTreeRegressor
from sklearn.neighbors import KNeighborsRegressor
from sklearn.neural_network import MLPRegressor
estimators = [LinearRegression()]
name_estimators = ["LinearRegression"]
balanced_accuracy_scorer = make_scorer(balanced_accuracy_score)
for j, est in enumerate(estimators):
results = {}
print("\n ---------- GenBooster +", name_estimators[j])
try:
for i, dataset in tqdm(enumerate(clf_datasets.items())):
dataset_name = dataset[0]
print("\n ----------", (i + 1), "/", len(clf_datasets.items()), ": ", dataset_name)
try:
X, y = dataset[1]['dataset'][0], dataset[1]['dataset'][1]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42, stratify=y)
X_train = X_train.astype(np.float64)
X_test = X_test.astype(np.float64)
# Split dataset into training and testing sets
model = BoosterClassifier(base_estimator=est)
model.fit(X_train, y_train)
y_pred_train = model.predict(X_train)
y_pred = model.predict(X_test)
# Calculate balanced accuracy
balanced_acc_train = balanced_accuracy_score(y_train, y_pred_train)
balanced_acc_test = balanced_accuracy_score(y_test, y_pred)
print("Training Balanced Accuracy:", balanced_acc_train)
print("Testing Balanced Accuracy:", balanced_acc_test)
results[dataset_name] = {"training_score": balanced_acc_train,
"testing_score": balanced_acc_test}
except Exception as e:
print("Error", e)
continue
except Exception as e:
print("Error", e)
continue
# Your data
data = results
# Convert to DataFrame - correct approach
df = pd.DataFrame.from_dict({(i,j): data[i][j]
for i in data.keys()
for j in data[i].keys()},
orient='index')
# The index is a MultiIndex with (dataset, score_type)
# Let's properly unpack it
df = df.reset_index()
df.columns = ['index', 'score'] # Temporary column names
# Split the multi-index into separate columns
df[['dataset', 'score_type']] = pd.DataFrame(df['index'].tolist(), index=df.index)
df = df.drop(columns=['index'])
# Now pivot the data
plot_df = df.pivot(index='dataset', columns='score_type', values='score')
# Sort by test score for better visualization
plot_df = plot_df.sort_values('testing_score')
# Create figure
plt.figure(figsize=(14, 10), dpi=100)
# Plot training and test scores
sns.scatterplot(data=plot_df, x=plot_df.index, y='training_score',
color='blue', label='Training Score', s=100, alpha=0.7)
sns.scatterplot(data=plot_df, x=plot_df.index, y='testing_score',
color='red', label='Test Score', s=100, alpha=0.7)
# Add connecting lines
for i, dataset in enumerate(plot_df.index):
plt.plot([i, i], [plot_df.loc[dataset, 'training_score'],
plot_df.loc[dataset, 'testing_score']],
color='gray', alpha=0.3, linestyle='--')
# Add perfect score line
plt.axhline(1.0, color='green', linestyle=':', alpha=0.5, label='Perfect Score')
# Customize plot
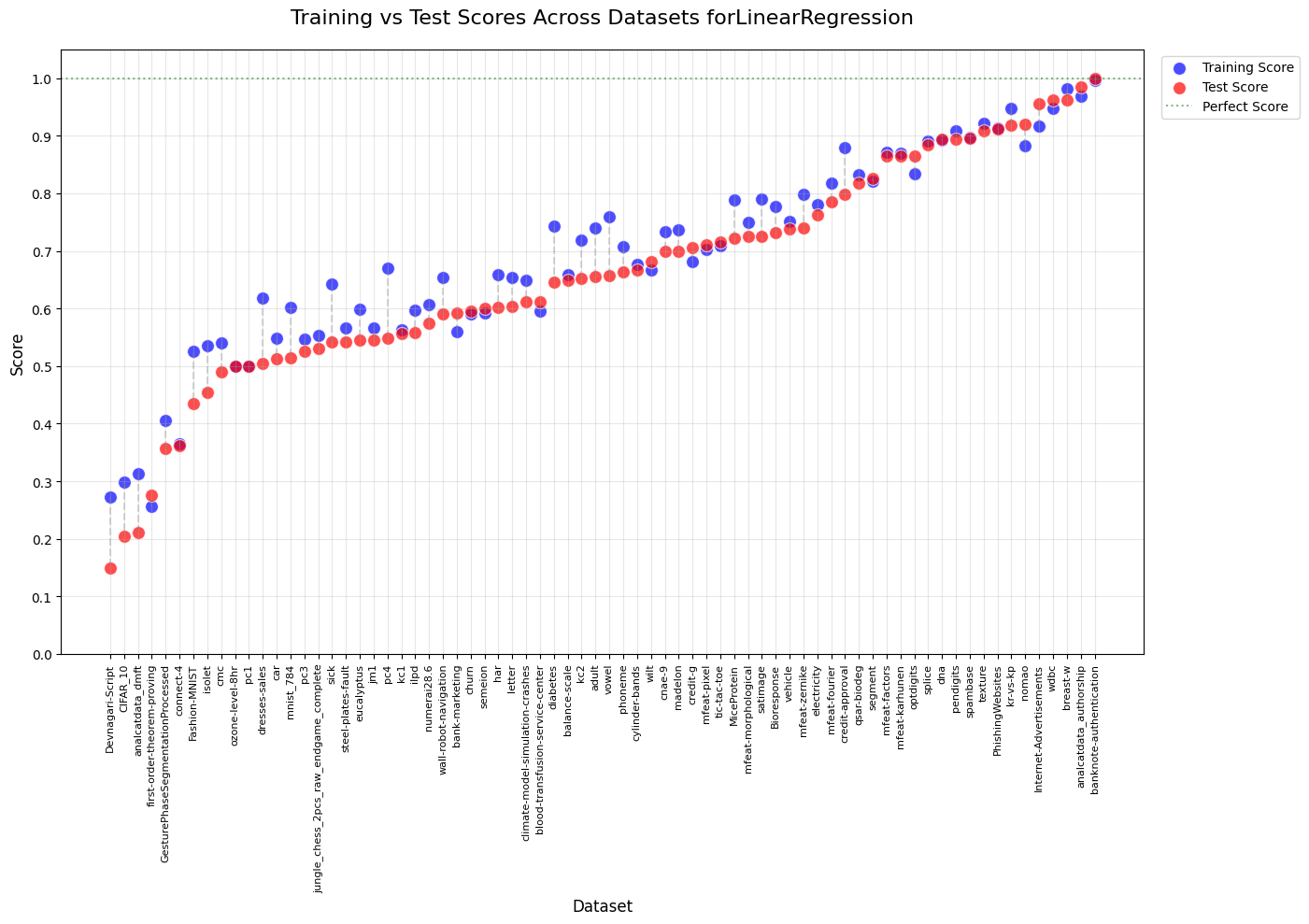
plt.title(f'Training vs Test Scores Across Datasets for' + name_estimators[j], fontsize=16, pad=20)
plt.xlabel('Dataset', fontsize=12)
plt.ylabel('Score', fontsize=12)
plt.xticks(rotation=90, fontsize=8)
plt.yticks(np.arange(0, 1.1, 0.1))
plt.ylim(0, 1.05)
plt.legend(loc='upper right', bbox_to_anchor=(1.15, 1))
plt.grid(True, alpha=0.3)
# Adjust layout
plt.tight_layout()
plt.show()
# Run the analysis
training_scores = [result['training_score'] for result in results.values()]
testing_scores = [result['testing_score'] for result in results.values()]
results = detect_overfitting(training_scores, testing_scores)
# Print results in a readable format
print("=== Overfitting Detection Report ===")
print(f"Datasets: {results['n_datasets']}")
print(f"Mean training score: {results['mean_train_score']:.3f}")
print(f"Mean test score: {results['mean_test_score']:.3f}")
print(f"Mean difference: {results['mean_difference']:.3f} (SD={results['std_difference']:.3f})")
print(f"Spearman rho: {stats.spearmanr(training_scores, testing_scores).correlation:.3f}")
print("\n=== Normality Test ===")
print(f"Shapiro-Wilk p-value: {results['normality_shapiro_p']:.4f}")
print("Interpretation:", "Normal distribution" if results['normal_distribution'] else "Non-normal distribution")
print("\n=== Statistical Tests ===")
print("1. Paired t-test:")
print(f" t = {results['paired_ttest']['statistic']:.3f}, p = {results['paired_ttest']['p_value']:.5f}")
print(f" Significant: {'✅' if results['paired_ttest']['significant'] else '❌'}")
print("\n2. Wilcoxon signed-rank test:")
print(f" W = {results['wilcoxon_test']['statistic']:.3f}, p = {results['wilcoxon_test']['p_value']:.5f}")
print(f" Significant: {'✅' if results['wilcoxon_test']['significant'] else '❌'}")
print("\n=== Effect Sizes ===")
print(f"Cohen's d: {results['cohens_d']:.3f} ({results['cohens_d_interpret']} effect)")
print(f"Rank-biserial correlation: {results['rank_biserial']:.3f} ({results['rank_biserial_interpret']} effect)")
---------- GenBooster + LinearRegression
0it [00:00, ?it/s]
---------- 1 / 72 : kr-vs-kp
1it [00:00, 2.21it/s]
Training Balanced Accuracy: 0.9480928346328172
Testing Balanced Accuracy: 0.9182957393483708
---------- 2 / 72 : letter
2it [00:08, 4.76s/it]
Training Balanced Accuracy: 0.6532232216708022
Testing Balanced Accuracy: 0.6037087912087912
---------- 3 / 72 : balance-scale
3it [00:08, 2.76s/it]
Training Balanced Accuracy: 0.6579835623313884
Testing Balanced Accuracy: 0.6492236337971365
---------- 4 / 72 : mfeat-factors
4it [00:10, 2.50s/it]
Training Balanced Accuracy: 0.8712500000000001
Testing Balanced Accuracy: 0.8649999999999999
---------- 5 / 72 : mfeat-fourier
5it [00:12, 2.35s/it]
Training Balanced Accuracy: 0.8174999999999999
Testing Balanced Accuracy: 0.7849999999999999
---------- 6 / 72 : breast-w
6it [00:13, 1.64s/it]
Training Balanced Accuracy: 0.9810158838019196
Testing Balanced Accuracy: 0.962409420289855
---------- 7 / 72 : mfeat-karhunen
7it [00:15, 1.80s/it]
Training Balanced Accuracy: 0.8699999999999999
Testing Balanced Accuracy: 0.865
---------- 8 / 72 : mfeat-morphological
8it [00:16, 1.70s/it]
Training Balanced Accuracy: 0.75
Testing Balanced Accuracy: 0.7250000000000001
---------- 9 / 72 : mfeat-zernike
9it [00:21, 2.61s/it]
Training Balanced Accuracy: 0.7987499999999998
Testing Balanced Accuracy: 0.74
---------- 10 / 72 : cmc
10it [00:21, 2.00s/it]
Training Balanced Accuracy: 0.539420682309971
Testing Balanced Accuracy: 0.48957420514548927
---------- 11 / 72 : optdigits
11it [00:23, 2.01s/it]
Training Balanced Accuracy: 0.8338839752605576
Testing Balanced Accuracy: 0.865
---------- 12 / 72 : credit-approval
12it [00:24, 1.49s/it]
Training Balanced Accuracy: 0.8800414474732983
Testing Balanced Accuracy: 0.7980625931445604
---------- 13 / 72 : credit-g
13it [00:24, 1.17s/it]
Training Balanced Accuracy: 0.6815476190476191
Testing Balanced Accuracy: 0.7059523809523809
---------- 14 / 72 : pendigits
14it [00:26, 1.46s/it]
Training Balanced Accuracy: 0.9082342205793651
Testing Balanced Accuracy: 0.8939849624060152
---------- 15 / 72 : diabetes
15it [00:27, 1.10s/it]
Training Balanced Accuracy: 0.7434696261682243
Testing Balanced Accuracy: 0.6457407407407407
---------- 16 / 72 : spambase
16it [00:27, 1.09it/s]
Training Balanced Accuracy: 0.8971008789190608
Testing Balanced Accuracy: 0.8953865467099069
---------- 17 / 72 : splice
17it [00:28, 1.19it/s]
Training Balanced Accuracy: 0.8904200133868808
Testing Balanced Accuracy: 0.8835470085470085
---------- 18 / 72 : tic-tac-toe
18it [00:28, 1.40it/s]
Training Balanced Accuracy: 0.7092305954129476
Testing Balanced Accuracy: 0.7153432835820895
---------- 19 / 72 : vehicle
19it [00:29, 1.33it/s]
Training Balanced Accuracy: 0.7512680047291669
Testing Balanced Accuracy: 0.7388257575757575
---------- 20 / 72 : electricity
20it [00:29, 1.54it/s]
Training Balanced Accuracy: 0.7804471490091439
Testing Balanced Accuracy: 0.7631713554987212
---------- 21 / 72 : satimage
21it [00:33, 1.45s/it]
Training Balanced Accuracy: 0.7904065610735302
Testing Balanced Accuracy: 0.7254887702816034
---------- 22 / 72 : eucalyptus
22it [00:34, 1.36s/it]
Training Balanced Accuracy: 0.5992921311574563
Testing Balanced Accuracy: 0.5444643212085073
---------- 23 / 72 : sick
23it [00:34, 1.09s/it]
Training Balanced Accuracy: 0.6421904761904762
Testing Balanced Accuracy: 0.5416666666666666
---------- 24 / 72 : vowel
24it [00:37, 1.45s/it]
Training Balanced Accuracy: 0.7588383838383838
Testing Balanced Accuracy: 0.6565656565656567
---------- 25 / 72 : isolet
25it [00:42, 2.61s/it]
Training Balanced Accuracy: 0.5353598014888339
Testing Balanced Accuracy: 0.4539835164835164
---------- 26 / 72 : analcatdata_authorship
26it [00:43, 2.07s/it]
Training Balanced Accuracy: 0.9691163839829222
Testing Balanced Accuracy: 0.9847136047215497
---------- 27 / 72 : analcatdata_dmft
27it [00:44, 1.79s/it]
Training Balanced Accuracy: 0.31318315240756367
Testing Balanced Accuracy: 0.2114058144165671
---------- 28 / 72 : mnist_784
28it [00:47, 2.33s/it]
Training Balanced Accuracy: 0.6018919115939201
Testing Balanced Accuracy: 0.5142492099814755
---------- 29 / 72 : pc4
29it [00:48, 1.77s/it]
Training Balanced Accuracy: 0.6706163207080265
Testing Balanced Accuracy: 0.5482954545454546
---------- 30 / 72 : pc3
30it [00:48, 1.37s/it]
Training Balanced Accuracy: 0.5466884375956731
Testing Balanced Accuracy: 0.525
---------- 31 / 72 : jm1
31it [00:49, 1.09s/it]
Training Balanced Accuracy: 0.5669233866908285
Testing Balanced Accuracy: 0.5454690237298934
---------- 32 / 72 : kc2
32it [00:49, 1.17it/s]
Training Balanced Accuracy: 0.7185861091424521
Testing Balanced Accuracy: 0.651697699890471
---------- 33 / 72 : kc1
33it [00:50, 1.36it/s]
Training Balanced Accuracy: 0.5635553470919324
Testing Balanced Accuracy: 0.5556403893872877
---------- 34 / 72 : pc1
34it [00:50, 1.52it/s]
Training Balanced Accuracy: 0.5
Testing Balanced Accuracy: 0.5
---------- 35 / 72 : adult
35it [00:50, 1.69it/s]
Training Balanced Accuracy: 0.7400971341967484
Testing Balanced Accuracy: 0.6551535087719298
---------- 36 / 72 : Bioresponse
36it [00:51, 1.84it/s]
Training Balanced Accuracy: 0.7767986723709284
Testing Balanced Accuracy: 0.732331888295191
---------- 37 / 72 : wdbc
37it [00:51, 2.11it/s]
Training Balanced Accuracy: 0.9478328173374613
Testing Balanced Accuracy: 0.9623015873015872
---------- 38 / 72 : phoneme
38it [00:52, 2.38it/s]
Training Balanced Accuracy: 0.707737690038575
Testing Balanced Accuracy: 0.6642024281764636
---------- 39 / 72 : qsar-biodeg
39it [00:52, 2.37it/s]
Training Balanced Accuracy: 0.8324861723727508
Testing Balanced Accuracy: 0.816967792615868
---------- 40 / 72 : wall-robot-navigation
40it [00:53, 1.80it/s]
Training Balanced Accuracy: 0.653627904631567
Testing Balanced Accuracy: 0.5904361071027737
---------- 41 / 72 : semeion
41it [00:55, 1.02s/it]
Training Balanced Accuracy: 0.5926607850658483
Testing Balanced Accuracy: 0.6
---------- 42 / 72 : ilpd
42it [00:55, 1.24it/s]
Training Balanced Accuracy: 0.597766939872203
Testing Balanced Accuracy: 0.5581148121899362
---------- 43 / 72 : madelon
43it [00:56, 1.44it/s]
Training Balanced Accuracy: 0.73625
Testing Balanced Accuracy: 0.7
---------- 44 / 72 : nomao
44it [00:56, 1.53it/s]
Training Balanced Accuracy: 0.8829423602789812
Testing Balanced Accuracy: 0.9193963930806036
---------- 45 / 72 : ozone-level-8hr
45it [00:58, 1.18it/s]
Training Balanced Accuracy: 0.5
Testing Balanced Accuracy: 0.5
---------- 46 / 72 : cnae-9
46it [01:00, 1.45s/it]
Training Balanced Accuracy: 0.7333304959709455
Testing Balanced Accuracy: 0.699604743083004
---------- 47 / 72 : first-order-theorem-proving
47it [01:02, 1.39s/it]
Training Balanced Accuracy: 0.2568945598032055
Testing Balanced Accuracy: 0.27648809523809526
---------- 48 / 72 : banknote-authentication
48it [01:02, 1.06s/it]
Training Balanced Accuracy: 0.9954954954954955
Testing Balanced Accuracy: 1.0
---------- 49 / 72 : blood-transfusion-service-center
49it [01:02, 1.22it/s]
Training Balanced Accuracy: 0.5951321966889054
Testing Balanced Accuracy: 0.611842105263158
---------- 50 / 72 : PhishingWebsites
50it [01:03, 1.41it/s]
Training Balanced Accuracy: 0.9133593601218816
Testing Balanced Accuracy: 0.9122886931875696
---------- 51 / 72 : cylinder-bands
51it [01:03, 1.72it/s]
Training Balanced Accuracy: 0.6772527472527472
Testing Balanced Accuracy: 0.6669004207573632
---------- 52 / 72 : bank-marketing
52it [01:03, 1.86it/s]
Training Balanced Accuracy: 0.5602668372475554
Testing Balanced Accuracy: 0.5917464996315401
---------- 53 / 72 : GesturePhaseSegmentationProcessed
53it [01:04, 1.44it/s]
Training Balanced Accuracy: 0.4052738438032556
Testing Balanced Accuracy: 0.3567099567099567
---------- 54 / 72 : har
54it [01:06, 1.17it/s]
Training Balanced Accuracy: 0.6586891684653934
Testing Balanced Accuracy: 0.6022327522327522
---------- 55 / 72 : dresses-sales
55it [01:06, 1.46it/s]
Training Balanced Accuracy: 0.6180213464696224
Testing Balanced Accuracy: 0.5049261083743842
---------- 56 / 72 : texture
56it [01:08, 1.16s/it]
Training Balanced Accuracy: 0.9217171717171717
Testing Balanced Accuracy: 0.9090909090909093
---------- 57 / 72 : connect-4
57it [01:09, 1.00s/it]
Training Balanced Accuracy: 0.36527957383567194
Testing Balanced Accuracy: 0.36229643372500514
---------- 58 / 72 : MiceProtein
58it [01:13, 2.01s/it]
Training Balanced Accuracy: 0.7886130536130537
Testing Balanced Accuracy: 0.7212312030075188
---------- 59 / 72 : steel-plates-fault
59it [01:15, 1.85s/it]
Training Balanced Accuracy: 0.5668243559177928
Testing Balanced Accuracy: 0.5421214137829045
---------- 60 / 72 : climate-model-simulation-crashes
60it [01:15, 1.38s/it]
Training Balanced Accuracy: 0.6486486486486487
Testing Balanced Accuracy: 0.6111111111111112
---------- 61 / 72 : wilt
61it [01:15, 1.06s/it]
Training Balanced Accuracy: 0.6660061646851607
Testing Balanced Accuracy: 0.6818181818181819
---------- 62 / 72 : car
62it [01:16, 1.08it/s]
Training Balanced Accuracy: 0.5475360592569449
Testing Balanced Accuracy: 0.5132936507936507
---------- 63 / 72 : segment
63it [01:17, 1.09s/it]
Training Balanced Accuracy: 0.821012708763058
Testing Balanced Accuracy: 0.8258268824771289
---------- 64 / 72 : mfeat-pixel
64it [01:23, 2.42s/it]
Training Balanced Accuracy: 0.7024999999999999
Testing Balanced Accuracy: 0.7100000000000001
---------- 65 / 72 : Fashion-MNIST
65it [01:27, 2.94s/it]
Training Balanced Accuracy: 0.5262499999999999
Testing Balanced Accuracy: 0.43499999999999994
---------- 66 / 72 : jungle_chess_2pcs_raw_endgame_complete
66it [01:27, 2.19s/it]
Training Balanced Accuracy: 0.5535725243283519
Testing Balanced Accuracy: 0.5305783752385694
---------- 67 / 72 : numerai28.6
67it [01:28, 1.67s/it]
Training Balanced Accuracy: 0.6070716881814764
Testing Balanced Accuracy: 0.5743074307430743
---------- 68 / 72 : Devnagari-Script
68it [01:38, 4.07s/it]
Training Balanced Accuracy: 0.2718190537084399
Testing Balanced Accuracy: 0.15
---------- 69 / 72 : CIFAR_10
69it [01:40, 3.60s/it]
Training Balanced Accuracy: 0.29875
Testing Balanced Accuracy: 0.205
---------- 70 / 72 : Internet-Advertisements
70it [01:41, 2.66s/it]
Training Balanced Accuracy: 0.9168827257490049
Testing Balanced Accuracy: 0.9555647840531561
---------- 71 / 72 : dna
71it [01:41, 2.07s/it]
Training Balanced Accuracy: 0.8917670682730924
Testing Balanced Accuracy: 0.8936965811965812
---------- 72 / 72 : churn
72it [01:42, 1.42s/it]
Training Balanced Accuracy: 0.5900049020872572
Testing Balanced Accuracy: 0.5955149501661129
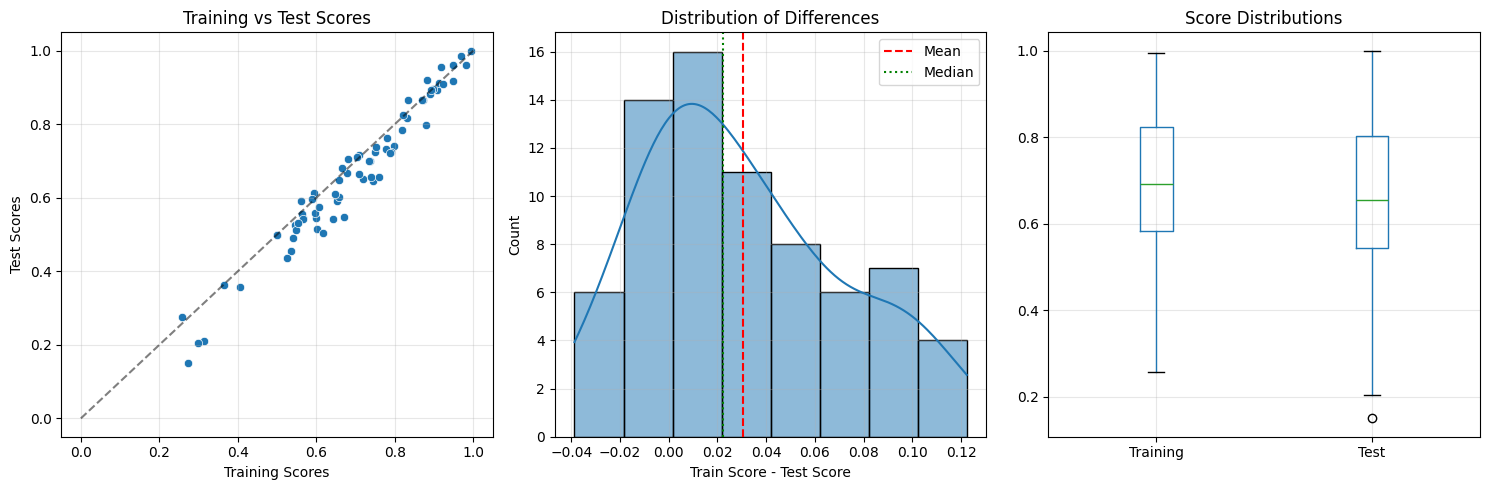
=== Overfitting Detection Report ===
Datasets: 72
Mean training score: 0.692
Mean test score: 0.661
Mean difference: 0.031 (SD=0.041)
Spearman rho: 0.972
=== Normality Test ===
Shapiro-Wilk p-value: 0.0153
Interpretation: Non-normal distribution
=== Statistical Tests ===
1. Paired t-test:
t = 6.302, p = 0.00000
Significant: ✅
2. Wilcoxon signed-rank test:
W = 2125.000, p = 0.00000
Significant: ✅
=== Effect Sizes ===
Cohen's d: 0.743 (medium effect)
Rank-biserial correlation: 0.191 (small effect)
But… Which one is the best in this particular setting?


Comments powered by Talkyard.